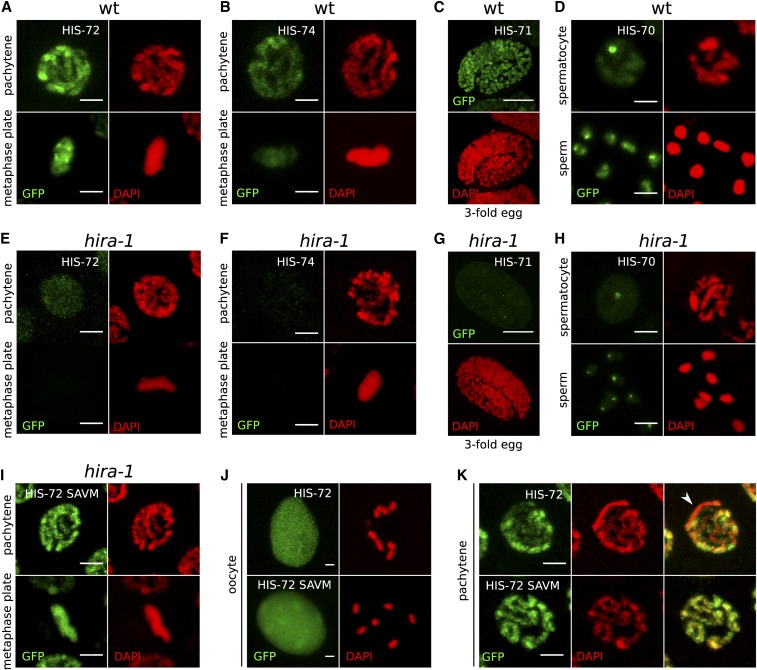
Figure 4.
Chromatin association of all H3.3 homologs depends on HIRA-1. (A–D) Chromatin association of H3.3 homologs in a wild-type (wt) background. (A) HIS-72 in pachytene and mitotic metaphase nuclei, (B) HIS-74 in pachytene and mitotic metaphase nuclei, (C) HIS-71 in threefold embryo, and (D) HIS-70 in spermatocyte nucleus and mature sperm. (E–H) H3.3 homolog signal is lost in hira-1 deletion background at the same stages as in (A–D). (E) HIS-72, (F) HIS-74, (G) HIS-71, and (H) HIS-70. Residual signal remains at HIS-70 foci. See text for details. (I–K) Chromatin localization of HIS-72 in hira-1 deletion background is restored upon mutation of the H3.3-specific motif in HIS-72 to the canonical H3-specific motif (AAIG to SAVM). (I) HIS-72 SAVM in pachytene and mitotic metaphase nuclei. (J and K) Chromatin association of HIS-72 (top panels) and HIS-72 SAVM (bottom panels) appears different. HIS-72 SAVM is more strongly chromatin associated in oocyte nuclei, with chromosomes being visible despite the strong nucleoplasmic background (J), and there is no obvious depletion of GFP signal in any part of the chromatin (K). Bars represent 2 µm in all panels except (C and G), where they represent 10 µm.