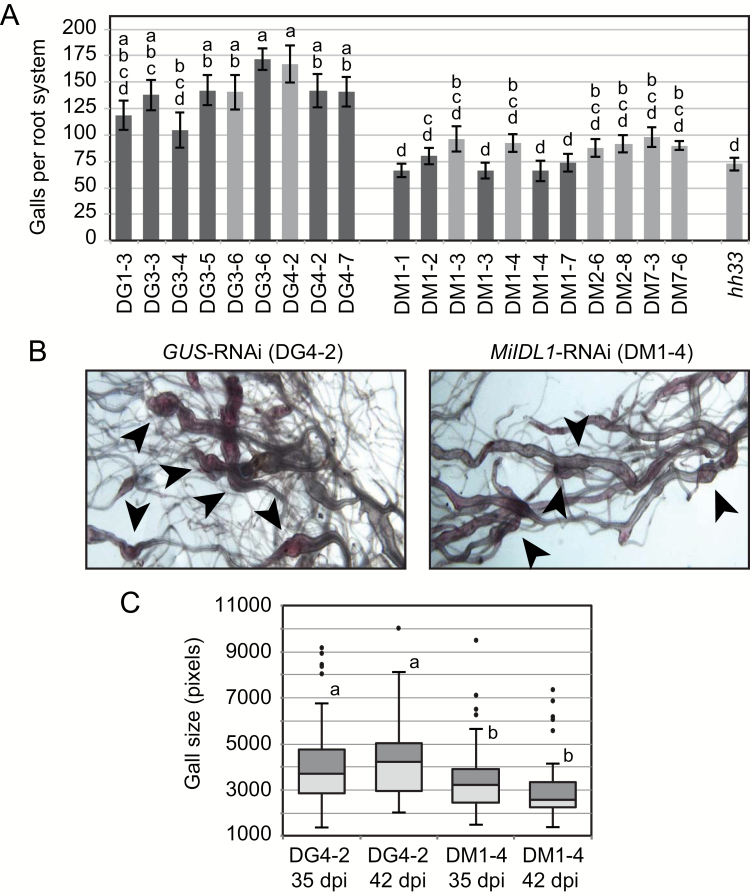
Fig. 5.
Gall number and size on M. incognita infected roots of wild-type Arabidopsis transformed with GUS and MiIDL1-RNAi. (A) Mean number of galls per root system of 10–12 independently grown plants for multiple transgenic lines for GUS (DG) and MiIDL1 (DM) at 35 dpi (light gray bars) and 42 dpi (darker gray bars). (B) Photos showing the relative size of galls at 35 dpi on GUS and MiIDL1-RNAi plants. (C) Box chart of gall sizes (pixels counted using ImageJ software) from multiple photos of different plants from the lines indicated (the area of 50–100 galls was measured per transgenic line). In (A), DG and DM indicate GUS or MiIDL1-RNAi, respectively, and the first number after DG or DM indicate the transformation event from T0 plants and the second number the seed collection (line) from T1 plants. hh33 is the hae-3/hsl2-3 double mutant. Galls were counted on T3 plants. The standard error bars in (A) reflect gall counts for 9–14 plants for each line. Letters above or next to the bars in (A, C) indicate means that are statistically grouped using a Tukey–Kramer test in SAS. The P value (two-tailed t-test) for the significance of the difference between the means for the GUS lines compared with the MiDL1 lines in (A) is 4.1 × 10−7. Arrows in (B) point to representative galls. Points in (C) are outliers.