The fungal jasmonate lasiojasmonate A functions as a source of jasmonic acid that activates the JA-isoleucine pathway in Arabidopsis.
Keywords: Jasmonate signalling pathway, jasmonic acid, Lasiodiplodia, lasiojasmonate A, phytotoxins
Abstract
Jasmonates are signaling compounds that regulate plant responses to stress. Jasmonic acid (JA) is the direct precursor of the bioactive plant hormone JA-isoleucine (JA-Ile), the ligand of the CORONATINE INSENSITIVE 1–jasmonate ZIM-domain (COI1–JAZ) co-receptor complex. JA, its methyl ester, and three furanonyl esters were recently isolated from the grapevine pathogen Lasiodiplodia mediterranea. The JA ester lasiojasmonate A (LasA) is the first reported naturally occurring JA-furanone, and its mode of action has not yet been elucidated. Here, we show that LasA activates many JA-regulated responses in planta, including protein degradation, gene expression, and physiological processes. These in vivo effects require LasA conversion into JA, formation of JA-Ile, and its recognition by the plant JA-Ile perception complex. These findings suggest a mode of action of the natural fungal LasA as an inactive JA pool that can be transformed into the bioactive JA-Ile form. We propose that fungal production of JA derivates such as LasA occurs at late infection stages to induce plant JA responses such as cell death, and can facilitate fungal infection.
Introduction
Fungi are producers of secondary metabolites that belong to different classes of natural compounds (Turner and Aldridge 1983; Cole et al., 2003; Lo Presti et al., 2015; Fonseca et al., 2018). Phytopathogenic fungi are among the main causative agents of disease in crops and forest plants, and lead to considerable economic losses (Evidente and Motta 2001; Evidente et al., 2010, 2011, 2013; Cimmino et al., 2014). For example, Botryosphaeriaceae is a widely spread family of pathogenic plant fungi associated with fruit rot, leaf spots, dieback, cankers, and root rot of angiosperms and gymnosperms (Phillips et al., 2013). Several species of the Botryosphaeriaceae, including Lasiodiplodia, infect grapevine worldwide (Larignon et al., 2001; Phillips, 2002; Van Niekerk et al., 2006; Pitt et al., 2010; Úrbez-Torres, 2011). In vitro, the newly discovered species Lasiodiplodia mediterranea (Linaldeddu et al., 2015) produces (1R,2R)-(-)-jasmonic acid (JA) as its main phytotoxin, the methyl ester (JA-Me), and three furanonenoyl esters, termed lasiojasmonates A, B, and C (LasA, B, C), as well as 16-O-acetylbotryosphaerilactones A and C, botryosphaerilactone A, (3S,4R,5R)-4-hydroxymethyl-3,5-dimethyldihydro-2-furanone, and (3R,4S)-botryodiplodin (Andolfi et al., 2014).
Jasmonic acid (JA) belongs to the naturally occurring family of compounds termed jasmonates (JAs); these are key plant signalling molecules that regulate several stress responses as well as developmental traits (Wasternack and Hause, 2013; Chini et al., 2016; Wasternack and Feussner, 2018). JA activates plant defenses against nematodes, herbivores, and necrotrophic pathogens (Wasternack and Hause, 2013; Goossens et al., 2016; Guo et al., 2018). JAs are cyclopentanone oxylipins derived from α-linolenic acid via lipid peroxidation (Schaller and Stintzi, 2009). The JA biosynthetic pathway and its intermediate compounds, such as 12-oxo-phytodienoic acid (OPDA), have been analysed extensively in plants (Schaller and Stintzi, 2009; Wasternack and Hause, 2013; Wasternack and Feussner, 2018; Wasternack and Song, 2017; Chini et al., 2018). In the final biosynthetic step of this pathway, the JA-amido synthetase JAR1 conjugates JA with isoleucine (Ile) to generate bioactive JA-Ile [(+)-7-iso-JA-Ile], which is in equilibrium with its inactive epimer (-)-JA-Ile (Staswick and Tiryaki, 2004; Fonseca et al., 2009). Bioactive JA-Ile acts as ‘molecular glue’ to promote formation of the COI1-JAZ (CORONATINE INSENSITIVE 1–jasmonate ZIM-domain) co-receptor complex (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Fonseca et al., 2009; Sheard et al., 2010). JA-Ile-induced COI1–JAZ interaction promotes ubiquitination and proteasome degradation of the JAZ repressors, which in turn activate several transcription factors that regulate specific physiological responses (Chini et al., 2007, 2016; Thines et al., 2007; Zhang et al., 2015; Wasternack and Feussner, 2018; Zhai et al., 2017).
Synthesis of phytohormones and phytohormone mimics is a common strategy of several pathogens to unbalance the plant immune system (Gimenez-Ibanez et al., 2016). The most-studied example is probably coronatine (COR), which is produced by some strains of Pseudomonas syringae. COR mimics the bioactive JA-Ile hormone by directly binding to the JA-Ile receptor COI1–JAZ (Brooks et al., 2005; Sheard et al., 2010). Lasiodiplodia mediterranea and other fungi also accumulate oxylipins such as OPDA, JA, and JA conjugates (Miersch et al., 1993; Andolfi et al., 2014; Chanclud and Morel, 2016; Fonseca et al., 2018). For example, Botryodiplodia theobromae, the causal agent of dieback disease of several plants, produces JA as a phytotoxin (Miersch et al., 1987; Gimenez-Ibanez et al., 2016). Fusarium oxysporum not only produces JA, but some strains also accumulate jasmonate conjugates such as JA-Ile and JA-Leu (Miersch et al., 1999; Brodhun et al., 2013; Cole et al., 2014). To promote infection, the activity of these jasmonates from F. oxysporum requires COI1, the plant JA-Ile receptor; this suggests a role for fungal JA-conjugates as virulent effectors (Cole et al., 2014). The rice blast fungus Magnaporthe oryzae was recently shown to convert natural JA into hydroxylated JA (12OH-JA), an inactive product of JA catabolism (Patkar et al., 2015). Although the activity of fungus-produced 12OH-JA is still not clear, secretion of this hydroxylated compound during infection to weaken plant defences suggests a role as a virulent factor (Patkar et al., 2015).
Although several JA derivatives have been isolated as fungal metabolites, their mode of action has not yet been determined. Here, we studied the effect of the natural fungal JA derivative LasA on plant JA-regulated responses, to define its mode of action in planta. LasA triggers many JA-regulated plant responses, including JAZ degradation, activation of JA gene expression, and growth inhibition. These effects in planta are dependent on JAR1 and COI1, which shows that plant conversion of LasA into JA, generation of JA-Ile, and its recognition by the JA-Ile receptor complex are necessary for the fungal LasA activity.
Materials and methods
Chemicals
Jasmonic acid and (1R,2R,3S,4R,5R)-4-hydroxymethyl-3,5-dimethyldihydro-2-furanone (LasA) were isolated from culture filtrates of Lasiodiplodia mediterranea (BL 101 strain), as reported by Andolfi et al. (2014). Coronatine was purchased from Sigma-Aldrich.
Plant material and growth conditions
Arabidopsis thaliana Col-0 was the genetic background of wild-type and mutant lines used in this study. Knock-out lines for coi1-1 (Xie et al., 1998), coi1-30 (Yang et al., 2012), coi1-2 (Xu et al., 2002), jar1-1 (Staswick and Tiryaki, 2004), myc2/jin1-2 (Lorenzo et al., 2004), and jai3-1 (Chini et al., 2007) have been described previously. Seeds were surface-sterilized by the chlorine gas method (in the presence of bleach and HCl for 3 h) and stratified for 2–3 d at 4 °C in the dark. All of the seedlings were grown under a 16-h light/8-h dark cycle at 21 °C (photosynthetic photon flux density approximately 100 µmol m–2 s–1). For root-growth inhibition assays, 10 to 30 seeds of each line were germinated for 10 d, with or without the presence of 50 µM jasmonic acid, 50 µM LasA, or 0.5 µM COR (Reveglia et al., 2018). Images were taken with a Nikon D1-x digital camera and root length was estimated using the ImageJ software (https://imagej.nih.gov/ij/). Data were analysed by one-way ANOVA/Tukey HSD post hoc tests. Four independent biological replicates (10–30 seedlings each) were measured for each sample with similar results. Experiments were repeated four times with similar results.
Anthocyanin measurement
Between 10 to 30 seeds of each line were germinated for 10 d, with or without the presence of 50 µM jasmonic acid, 50 µM LasA, or 0.5 µM COR. Between four to ten seedlings were weighed and pooled for each biological replicate; three independent pools were measured for each line. Seedlings were soaked overnight in 1 ml of hydrochloric acid in aqueous methanol (HCl 0.5 N; methanol 80%, v/v). The reference blank was obtained by adding half of the sample solution to a peroxide reagent (one part 30% hydrogen peroxide in nine parts of methanolic hydrochloric acid, 5:1 v/v, 3N), whereas the other half of the solution was diluted with one volume of methanolic hydrochloric acid (5:1 v/v, 3N). Absorbance was measured at 530 nm, and anthocyanin content was expressed as (A530 sample – A530 blank) per g fresh weight. Data were analysed by one-way ANOVA/Tukey HSD post hoc tests. Experiments were repeated twice with similar results.
Yeast two-hybrid system
All constructs were previously reported by Chini et al. (2007). pGAD (prey) and pGBK (bait) plasmids were co-transformed in Saccharomyces cerevisiae AH109 cells using standard heat-shock protocols (Chini et al., 2007). Successfully transformed colonies were selected 3 d after transformation on yeast synthetic drop-out medium lacking Leu and Trp (–2 medium). Yeast colonies were then grown in selective –2 liquid medium for ~5 h and cell density adjusted to 3 × 107 cells ml–1 (OD600=1). Cell suspensions (5 µl) were plated on yeast synthetic drop-out medium lacking Ade, His, Leu, and Trp (–4 medium) to evaluate protein interactions. Where indicated, the medium was supplemented with 20 µM coronatine and/or 100 µM LasA. Plates were incubated (2–6 d; 28 °C) and images were acquired with a Nikon D1-x digital camera.
JAZ1-GUS staining and quantification assays
The 35S:JAZ1-GUS in the wild-type and coi1-30 backgrounds have been described previously (Thines et al., 2007; Monte et al., 2014). The 35S:JAZ1-GUS marker line was introgressed into the jar1-1 background by crossing, and a double-homozygous line was used for further analyses. 35S:JAZ1-GUS seedlings were grown vertically on Murashige and Skoog (MS) plates, whereas 35S:JAZ1-GUS coi1-30 seedlings were selected vertically on MS plates in the presence of 0.5 µM COR, and 6-d-old seedlings were treated for 1 h with either liquid Johnson medium (control), 5 µM JA, 100 µM LasA, or 1 µM COR solution, as described previously (Chini, 2014; Reveglia et al., 2018). Samples were then placed in standard GUS staining solution [50 mM phosphate buffer, pH 7; 0.5 mM K4Fe(CN)6; 0.5 mM K3Fe(CN)6 0.2% Triton X-100; 0.7 mg ml–1 5-bromo-4-chloro-3-indolyl β-D-glucuronic acid (X-Gluc)] and incubated overnight at 37 °C (Fernández-Calvo et al., 2011) to visualize GUS activity. Root images were acquired with a Nikon D1-x camera. The analysis was performed using 10–30 plants per sample; the experiment was repeated three times with similar results. For quantification of JAZ1-GUS protein degradation, approximately 50 seedlings were treated as described above, and the roots were collected and frozen in liquid nitrogen as previously described (Monte et al., 2014). The roots were homogenized in extraction buffer containing 50 mM phosphate buffer, pH 7, 10 µM β-mercaptoethanol, 10 mM EDTA, 0.1% sarcosyl (N-lauroylsarcosine sodium salt), and 0.1% Triton X-100. The total protein content was quantified by the Bradford method (Bio Rad Protein Assay). Then, a 30-μl extract was incubated with 70 μl protein extraction buffer containing 1 mM MUG (methylumbelliferyl-β-D-glucuronide hydrate) for 1 h at 37 °C, and 10-μl samples were taken at t=0 and t=1 h. Finally, 90 μl of 0.2 M Na2CO3 was used to stop the reaction. Fluorescence was measured at 365/460 nm (excitation/emission) with a SpectraMax M2 spectrophotometer (Molecular Devices). Six independent replicates were measured.
pJAZ2-GUS expression assays
The pJAZ2:GUS marker line was described previously by Gimenez-Ibanez et al. (2017). For pJAZ2:GUS expression experiments, seeds were germinated as described above and 6-d-old seedlings were treated with either liquid Johnson medium (control), 5 µM JA, or 100 µM LasA (2 h); GUS was stained as described above and incubated overnight at 37 ºC. Images were acquired with a Nikon D1-x camera. Analyses were performed using 10–20 plants per sample and the experiment was repeated three times with similar results.
Quantitative RT-PCR
Quantitative RT-PCR was performed using biological samples of tissue pooled from 10–20 wild-type seedlings. RNA was extracted and purified using Trizol reagent (Invitrogen) followed by use of a High Pure RNA isolation kit (Roche), including DNase digestion to remove genomic DNA contamination. cDNA was synthesized from 1 μg total RNA with a high-capacity cDNA reverse transcription kit (Applied Biosystems). For gene amplification, 4 μl from a 1:10 cDNA dilution was added to 4 μl of EvaGreen® qPCR Mix Plus (Solis BioDyne) and gene-specific primers as previously described (Chini et al., 2018). Quantitative PCR was performed in 96-well optical plates in a HT 7900 Real Time PCR system (Applied Biosystems) using standard thermocycler conditions (an initial hold at 95 °C for 10 min, followed by a two-step SYBRPCR program of 95 °C for 15 s and 60 °C for 60 s for 40 cycles). Relative expression values given as the means of three or four technical replicates relative to the basal wild-type control using ACT8 as the housekeeping gene. Data were analysed using unpaired Student’s t-tests.
Results
Effect of LasA in planta
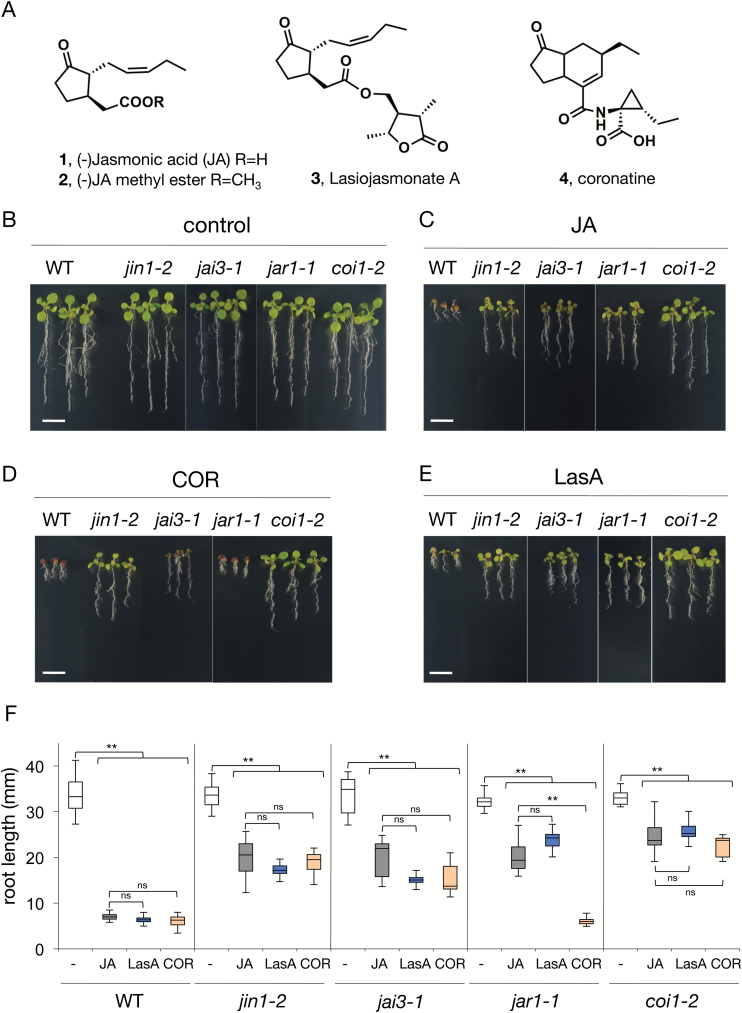
(–)-JA, its methyl ester, and lasiojasmonate A (Fig. 1A, 1–3) were purified from the organic extracts of Lasidiplodia mediterranea (BL 101 strain) culture filtrates and identified by comparing their physical and spectroscopic properties with those previously reported (Andolfi et al., 2014). To assess its effects in planta, wild-type (WT) Arabidopsis plants were grown with or without fungal LasA. Similar to JA, LasA induced growth inhibition and anthocyanin accumulation (Fig. 1B–F; Supplementary Fig. S1 at JXB online). To test whether the activity of this JA derivative required in planta conjugation of JA to Ile, JA-Ile perception, and the JA signalling pathway, we also grew seeds of jar1-1 (impaired in JA-Ile conjugation), coi1-2 (weak allele of the JA-Ile receptor), and jai3-1 and jin1-2 (both JA signalling mutants) in the presence of LasA. LasA showed an effect similar to that of JA in all genotypes, strongly inhibiting growth of WT plants but only partially inhibiting jar1-1, jai3-1, jin1-2, and coi1-2 (Fig. 1B–F; Supplementary Table S1). LasA induced accumulation of the defence secondary metabolites anthocyanins, and this response required a functional JA pathway (Supplementary Fig. S1).
Fig. 1.
Effects of LasA on Arabidopsis plants. (A) Structure of jasmonic acid (1), methyl jasmonate (2), lasiojasmonate A (3), and COR (4). (B–F) Wild-type (Col-0) and mutant seedlings (n=8–25) were germinated in the absence (control, B) or presence of 50 µM JA (C), 50 µM LasA (D), or 0.5 µM COR (E). Scale bars are 1 cm. (F) Root length was measured 10 d after germination. Data are shown as box-plots (control in white boxes, treatments with JA, LasA, and COR in grey, blue, and orange, respectively); horizontal lines are medians, the boxes show the upper and lower quartiles, and the whiskers show the full data range. Asterisks indicate significant differences compared to the untreated control or JA treatment for each plant genotype, evaluated by one-way ANOVA/Tukey HSD post hoc tests (** P<0.01). Experiments were repeated four times with similar results.
These results show that fungal LasA triggers JA-regulated responses, and that this LasA-induced effect requires the canonical JA pathway.
Effect of LasA on JA-mediated plant transcriptional activation
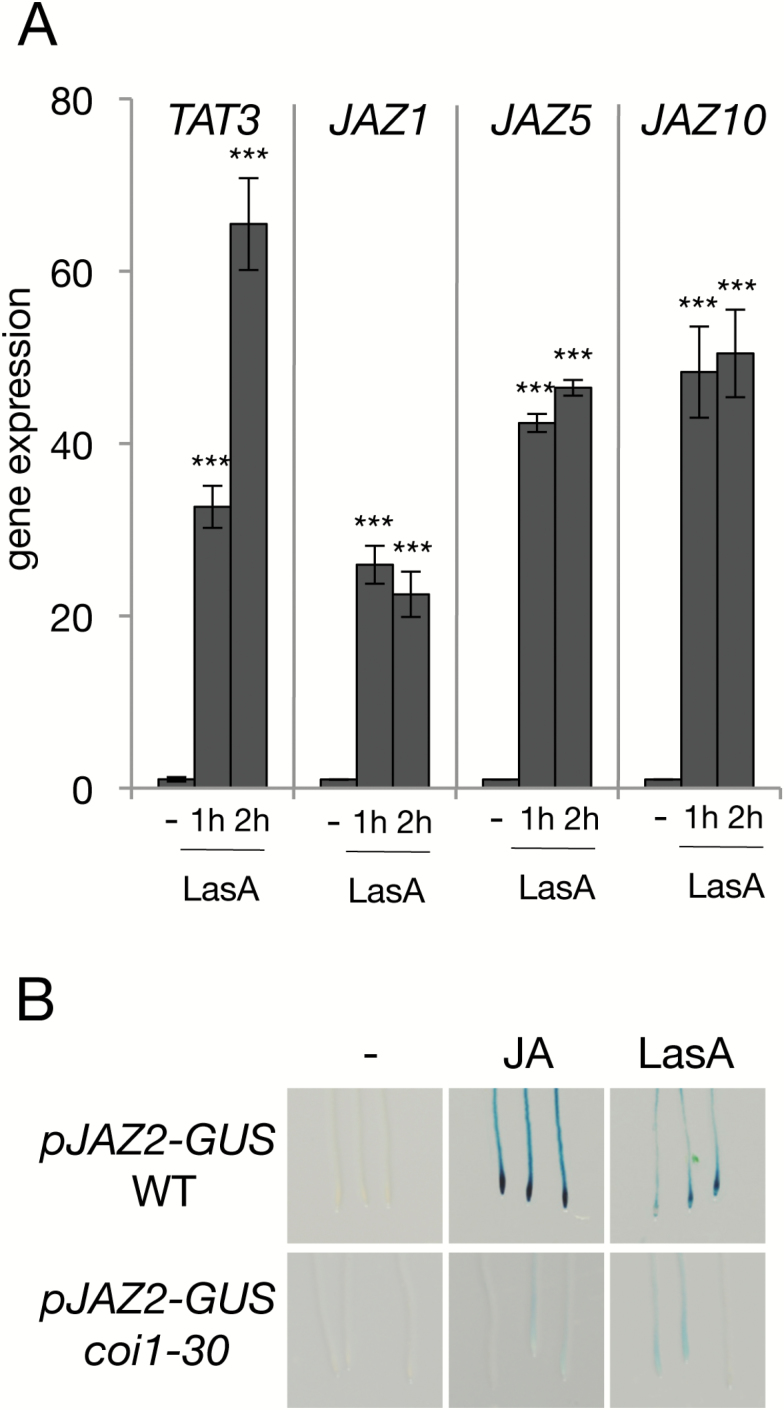
As LasA triggers physiological responses mediated by JA, we evaluated whether it could activate JA-dependent gene expression. We analysed the transcriptional changes of well-known JA-marker genes induced by LasA, including several JAZ genes as well as JA-biosynthetic genes such as LOX3, AOS, AOC1, and OPR3. Exogenous treatment of LasA triggered the expression of all tested JA-marker genes within 1 h (Fig. 2A; Supplementary Fig. S2).
Fig. 2.
Effects of LasA on JA-regulated transcriptional activation. (A) Gene expression analysis of TAT3, JAZ1, JAZ5, and JAZ10 in wild-type (Col-0) plants in response to 50 µM LasA for 1 or 2 h; untreated plants (–) were included as controls. ACT8 was used as the housekeeping control gene. Statistically significant differences in gene expression between controls and LasA-treated plants were determined by two-tailed Student’s t-test (*** P<0.001). Each biological sample consisted of tissue pooled from 10–15 plants. Data are means (±SD) of three or four technical replicates. (B) GUS-staining for visualization of pJAZ2-GUS expression in roots of transgenic Arabidopsis in the wild-type and coi1-30 backgrounds. Seedlings (7-d-old) were treated with 5 µM JA or 100 µM LasA; untreated plants (–) were included as controls.
To provide further evidence of the role of LasA in the activation of JA signalling, we assessed the effect of LasA on the JA-inducible marker line pJAZ2:GUS. As expected, LasA activated expression of the marker line, although at a lower level than JA (Fig. 2B). To assess whether this activation required JA-Ile perception by COI1, we tested activation of the marker line in the coi1-30 background. Similar to JA, fungal JA derivatives did not activate pJAZ2:GUS expression in coi1-30 mutant plants (Fig. 2B).
These data indicate that LasA induces expression of JA-regulated genes in a COI1-dependent manner.
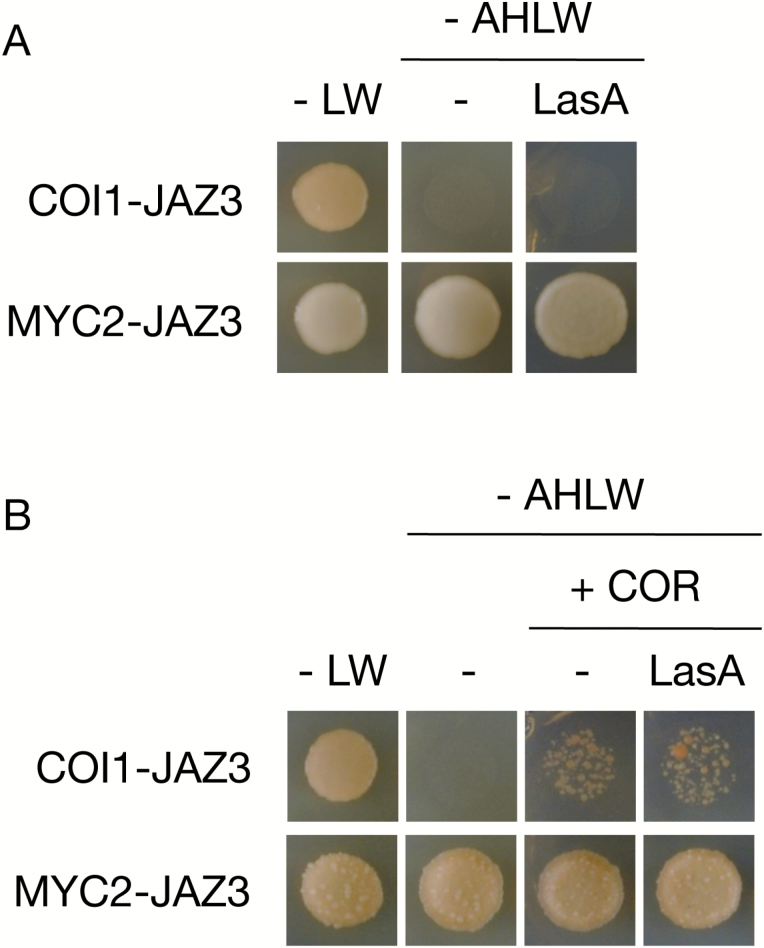
Effect of LasA on the JA-Ile co-receptor COI1–JAZ complex
LasA triggers physiological responses mediated by JA in a COI1- and JAR1-dependent manner (Fig. 1), suggesting that it is first converted to JA, then conjugated to form JA-Ile that would in turn trigger JAZ protein degradation. To support this hypothesis, we evaluated the LasA effect on hormone-mediated formation of the COI1–JAZ co-receptor complex in a yeast-two-hybrid assay (Y2H; Chini et al., 2009; Chini, 2014). LasA did not promote COI1–JAZ3 interaction in the absence of the hormone (Fig. 3A). To assess a possible LasA effect on yeast growth and Y2H detection of protein–protein interactions, we also assessed LasA activity on formation of the complex between JAZ3 and the transcription factor MYC2. LasA did not alter the JAZ3–MYC2 interaction. In the presence of COR, COI1 binds to JAZ3; LasA nonetheless did not affect the COR-induced COI1–JAZ3 interaction (Fig. 3B). These findings show that LasA does not directly interfere with the JA-Ile receptor complex COI1–JAZ.
Fig. 3.
Effects of LasA on the JA-Ile receptor COI1–JAZ. (A, B) Yeast cells co-transformed with pGAD-JAZ3 (prey) and pGBK-COI1 (bait) were selected and subsequently grown on synthetic drop-out medium lacking Leu and Trp (–LW) as the transformation control, or on selective media lacking Ade, His, Leu, and Trp (–AHLW) to test protein interactions. COI1 interaction with JAZ3 was detected only in the presence 20 µM coronatine (B). LasA (at 100 µM) did not alter COI1/JAZ interactions in the presence of COR. As a control, the interaction between JAZ3 (bait) and MYC2 (prey) was also assessed (A, lower panels). LasA did not promote COI1/JAZ3 interaction, nor alter the MYC2/JAZ3 interaction (B, lower panels).
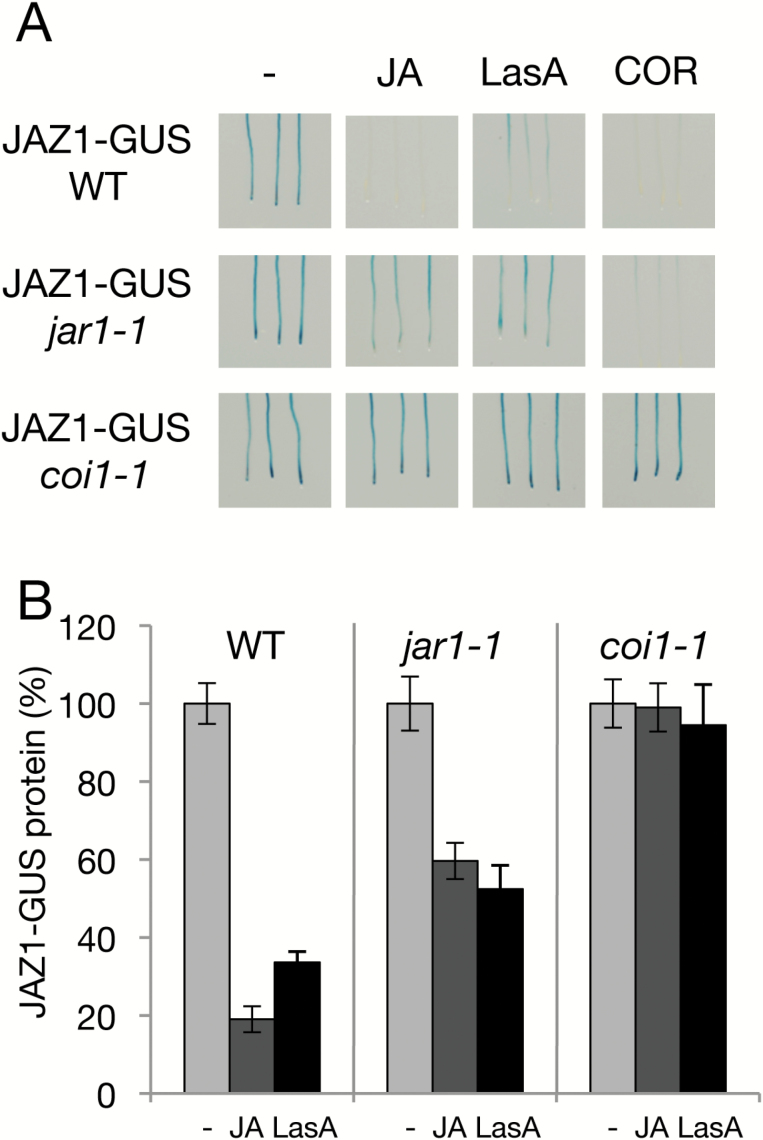
Effect of LasA on JAZ turnover
LasA induced JA-dependent gene expression without interfering directly with the COI1–JAZ receptor; therefore, LasA might be converted to JA and then conjugated to form JA-Ile, which in turn would trigger JAZ protein degradation. To test this hypothesis, we evaluated the effect of LasA on JAZ repressor degradation by studying JAZ1-GUS turnover in the presence of LasA. Similar to JA or COR treatments, LasA induced JAZ1 degradation (Fig. 4A). To determine whether JA must be conjugated into JA-Ile for LasA activity, we assessed the effects of LasA on JAZ1 stability in jar1-1 mutants, which have partially impaired JA-Ile conjugation. LasA induced a partial JAZ1-GUS degradation, similar to that promoted by JA (Fig. 4A, B). In addition, we studied the effect of LasA in coi1-1 plants that have loss of JA-Ile receptor function, in order to verify the requirement for JA-Ile perception for LasA activity. LasA did not induce JAZ1-GUS degradation in the complete loss-of-function coi1-1 mutant (Fig. 4A, B) and, as expected, JA also did not promote JAZ1-GUS degradation in this mutant. The JA-Ile analogue COR acts as a ligand of the COI1-JAZ receptor and activates JAZ degradation and JA responses independently of JAR1 (Sheard et al., 2010) and indeed, our results showed that COR induced JAZ1 degradation in a COI1-dependent, JAR1-independent manner (Fig. 4A). Finally, LasA did not prevent hormone-triggered degradation of JAZ1 during concurrent JA and LasA treatment (Supplementary Fig. S3).
Fig. 4.
Effects of LasA on JAZ stability. GUS visualization (A) and quantification of GUS activity (B) of JAZ1-GUS in roots of 7-d-old transgenic Arabidopsis 35S:JAZ1-GUS in the wild-type and mutant backgrounds. 35S:JAZ1-GUS wild-type and mutant plants (n=20) were treated with 5 µM JA, 100 µM LasA, or 1 µM COR for 1h (A). Untreated control plants (–) are also shown. Experiments were repeated twice with similar results. For JAZ1-GUS quantification, Arabidopsis 35S:JAZ1-GUS in the wild-type and mutant backgrounds (n=15–30) were exposed to 25 µM JA or 50 µM LasA for 1 h and GUS activity was measured in the roots (B). Untreated control plants (–) are also shown. Data are the means (±SD) of six replicates.
These results show that LasA induces JAZ degradation, and that its activity requires JAR1-mediated JA conversion into JA-Ile, as well as COI1-dependent perception of JA-Ile.
Discussion
Jasmonates are signalling molecules that govern plant responses to stress. Jasmonic acid is the direct precursor of the bioactive plant hormone JA-Ile, the endogenous ligand of the plant receptor complex. Several fungi produce JA and JA derivates; phytotoxic activity is reported for most of these compounds, although a precise molecular mode of action has yet to be proposed. Here, we addressed the activity of the natural JA derivate LasA, which is produced by the pathogenic fungus Lasiodiplodia mediterranea (Andolfi et al., 2014). Our results showed that LasA activated the plant JA pathway and that its activity required JAR1 and COI1, key proteins required for JA conversion into JA-Ile and for JA-Ile perception, respectively. Because LasA is a fungal JA furanonyl ester, we propose that this naturally occurring fungal compound can be catabolized to release JA, which is in turn converted into JA-Ile by JAR1. In support of this hypothesis, LasA activity required JAR1 for JA conjugation to Ile, as well as recognition by the JA-Ile perception complex. LasA might therefore function as an inactive JA conjugate that can be hydrolysed to liberate the direct JA-Ile precursor JA.
A cleaving amidohydrolase activity on JA conjugates has been described for the JA-producing fungus Botryodiplodia theobromae (Hertel et al., 1997). This enzymatic activity might be conserved among fungi, which would explain the detection of notable amounts of JA in several species with only minimal accumulation of JA-conjugated molecules (Chanclud and Morel, 2016; Lo Presti et al., 2015; Eng et al., 2016; Fonseca et al., 2018); however, the fungal cleaving amidohydrolase activity on JA conjugates might mask identification of additional natural JA-conjugates under in vitro conditions. Plants have also evolved amidohydrolase activity; the plant enzyme IAR3/ILL6 deconjugates JA-Ile into JA (Woldemariam et al., 2012; Bhosale et al., 2013; Widemann et al., 2013). This process might accept different substrates, since IAR3/ILL6 can also deconjugate 12OH–JA-Ile into 12OH-JA (Widemann et al., 2013). In addition, several members of this amidohydrolase family release free auxin (IAA) by cleaving IAA–amino acid conjugates, with each hydrolase deconjugating a different subset of the conjugates (LeClere et al., 2002). Finally, the expression of several amidohydrolase genes is induced by auxins and jasmonate, strengthening the case for the involvement of amidohydrolase in hormone signalling (LeClere et al., 2002; Zhang et al., 2016). Cleaving amidohydrolase activity of molecule–amino acid conjugates is thus a fairly common mechanism in nature, and both fungi and plants might be able to hydrolyse JA-conjugates such as LasA.
LasA activation of JA responses is dependent on the JA-Ile co-receptor COI1. A similar requirement for the plant COI1 JA-Ile receptor was reported for the virulent activity of fungal JA-conjugates produced by Fusarium oxysporum (Miersch et al., 1999; Brodhun et al., 2013; Cole et al., 2014). These data suggest that different fungi have evolved distinct JA-conjugates with similar biological activity dependent on the plant COI1 JA-Ile receptor.
Jasmonic acid regulates several plant responses, including plant defences against necrotroph pathogens, fungi, and herbivores (Wasternack and Hause, 2013; Goossens et al., 2016; Guo et al., 2018). On the other hand, activation of the JA pathway weakens the defences against biotrophic and hemibiotrophic pathogens triggered by salicylic acid (SA) (Robert-Seilaniantz et al., 2011). Hence, some strains of Pseudomonas syringae produce a mimic of the bioactive JA-Ile hormone, coronatine, to activate the JA pathway, which in turn inhibits the SA-dependent defences required for P. syringae resistance (Cui et al., 2005; Robert-Seilaniantz et al., 2011). A biotrophic or hemibiotrophic infection stage for L. mediterranea has not been reported to date, providing a case against the hypothesis that L. mediterranea produces LasA to activate JA-dependent responses to inhibit SA defences.
The suggested mode of action for fungal LasA—that it functions as a pool of inactive conjugated JA that is converted to active JA, and hence activates plant JA responses including defence against necrotrophic fungi—might seem counterintuitive at first glance. However, necrotrophic fungi induce plant cell death, which is beneficial for fungal growth and proliferation (Chanclud and Morel, 2016; Lo Presti et al., 2015). We therefore propose that fungal production of LasA, and possibly of additional JA derivates, would be spatio-temporally regulated to activate JA-mediated senescence and cell death only in specific conditions. For example, to avoid inducing plant defences, fungi would not produce LasA in early infection stages, whereas production and subsequent induction of plant cell death would occur in late stages in order to facilitate fungal propagation and infection. Future analysis of Lasiodiplodia mutants with impaired LasA production will test this hypothesis.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Accumulation of anthocyanins in response to exogenous treatment with JA, LasA, or COR.
Fig. S2. Effects of LasA on JA-regulated transcriptional activation.
Fig. S3. Effect of LasA on JA-mediated JAZ degradation.
Table S1. Data for root growth measurements reported in Fig. 1F.
Acknowledgements
We thank C. Mark for editorial assistance and L. Maddau (Dipartimento di Agraria, Sezione di Patologia Vegetale ed Entomologia, Università degli Studi di Sassari, Sassari, Italy) for the production of L. mediterranea culture filtrates. A. Evidente is associated with the Istituto di Chimica Biomolecolare del CNR, Pozzuoli, Italy. Work in the lab of AC and RS was funded by the Spanish Ministry for Science and Innovation (MINECO-AEI/FEDER grant BIO2016-77216-R) and CSIC (201740I018).
References
- Andolfi A, Maddau L, Cimmino A, Linaldeddu BT, Basso S, Deidda A, Serra S, Evidente A. 2014. Lasiojasmonates A–C, three jasmonic acid esters produced by Lasiodiplodia sp., a grapevine pathogen. Phytochemistry 103, 145–153. [DOI] [PubMed] [Google Scholar]
- Bhosale R, Jewell JB, Hollunder J, et al. . 2013. Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants. The Plant Cell 25, 2865–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodhun F, Cristobal-Sarramian A, Zabel S, Newie J, Hamberg M, Feussner I. 2013. An iron 13S-lipoxygenase with an α-linolenic acid specific hydroperoxidase activity from Fusarium oxysporum. PLoS ONE 8, e64919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, Kunkel BN. 2005. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Molecular Plant Pathology 6, 629–639. [DOI] [PubMed] [Google Scholar]
- Chanclud E, Morel JB. 2016. Plant hormones: a fungal point of view. Molecular Plant Pathology 17, 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A. 2014. Application of yeast-two hybrid assay to chemical genomic screens: a high-throughput system to identify novel molecules modulating plant hormone receptor complexes. Methods in Molecular Biology 1056, 35–43. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. 2009. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. The Plant Journal 59, 77–87. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, et al. . 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R. 2016. Redundancy and specificity in jasmonate signalling. Current Opinion in Plant Biology 33, 147–156. [DOI] [PubMed] [Google Scholar]
- Chini A, Monte I, Zamarreño AM, et al. . 2018. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nature Chemical Biology 14, 171–178. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Andolfi A, Evidente A. 2014. Phytotoxic terpenes produced by phytopathogenic fungi and allelopathic plants. Natural Product Communications 9, 401–408. [PubMed] [Google Scholar]
- Cole RJ, Jarvis BB, Schweikert MA. 2003. Handbook of secondary fungal metabolites, vols 1–3 London: Academic Press. [Google Scholar]
- Cole SJ, Yoon AJ, Faull KF, Diener AC. 2014. Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Molecular Plant Pathology 15, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. 2005. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proceedings of the National Academy of Sciences, USA 102, 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng F, Haroth S, Feussner K, Meldau D, Rekhter D, Ischebeck T, Brodhun F, Feussner I. 2016. Optimized jasmonic acid production by Lasiodiplodia theobromae reveals formation of valuable plant secondary metabolites. Plos ONE 11, e0167627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evidente A, Andolfi A, Cimmino A. 2011. Relationships between the stereochemistry and biological activity of fungal phytotoxins. Chirality 23, 674–693. [DOI] [PubMed] [Google Scholar]
- Evidente A, Andolfi A, Cimmino A, Abouzeid MA. 2010. Phytotoxins produced by fungi responsible of forest plant diseases. In: Salaazr A, Rios I, eds. Sustainable agriculture – technology, planning and management. New York: Nova Science Publishers, Inc, 177–234. [Google Scholar]
- Evidente A, Cimmino A, Andolfi A. 2013. The effect of stereochemistry on the biological activity of natural phytotoxins, fungicides, insecticides and herbicides. Chirality 25, 59–78. [DOI] [PubMed] [Google Scholar]
- Evidente A, Motta A. 2001. Phytotoxins from fungi, pathogenic for agrarian, forest and weedy plants. In: Tringali C, ed. Bioactive compounds from natural sources: isolation, characterization and biological properties. London: Taylor and Francis, 473–526. [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. . 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. 2009. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nature Chemical Biology 5, 344–350. [DOI] [PubMed] [Google Scholar]
- Fonseca S, Radhakrishnan D, Prasad K, Chini A. 2018. Fungal production and manipulation of plant hormones. Current Medicinal Chemistry 25, 253–267. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Ortigosa A, García-Casado G, Chini A, Lewsey MG, Ecker JR, Ntoukakis V, Solano R. 2017. JAZ2 controls stomata dynamics during bacterial invasion. New Phytologist 213, 1378–1392. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Chini A, Solano R. 2016. How microbes twist jasmonate signaling around their little fingers. Plants 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens J, Fernández-Calvo P, Schweizer F, Goossens A. 2016. Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Molecular Biology 91, 673–689. [DOI] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA. 2018. Resolution of growth–defense conflict: mechanistic insights from jasmonate signaling. Current Opinion in Plant Biology 44, 72.– . [DOI] [PubMed]
- Hertel SC, Knöfel HD, Kramell R, Miersch O. 1997. Partial purification and characterization of a jasmonic acid conjugate cleaving amidohydrolase from the fungus Botryodiplodia theobromae. FEBS Letters 407, 105–110. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. 2008. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proceedings of the National Academy of Sciences, USA 105, 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larignon P, Fulchic R, Cere L, Dubo B. 2001. Observation on black dead arm in French vineyards. Phytopathologia Mediterranea 40, 336–342. [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B. 2002. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. The Journal of Biological Chemistry 277, 20446–20452. [DOI] [PubMed] [Google Scholar]
- Linaldeddu BT, Deidda A, Scanu B, Franceschini A, Serra S, Berraf-Tebbal A, Bouiti MZ, Ben Jamâa ML, Phillips AJL. 2015. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Diversity 71, 201–214. [Google Scholar]
- Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R. 2015. Fungal effectors and plant susceptibility. Annual Review of Plant Biology 66, 513–545. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O, Bohlmann H, Wasternack C. 1999. Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry 50, 517–523. [Google Scholar]
- Miersch O, Gunther T, Fritsche W, Sembdner G. 1993. Jasmonates from different fungal species. Natural Product Letters 2, 293–299. [Google Scholar]
- Miersch O, Preiss K, Schreiber GS. 1987. (+)-7-iso-Jasmonic acid and related compounds from Botryodiplodia theobromae. Phytochemistry 26, 1037–1039. [Google Scholar]
- Monte I, Hamberg M, Chini A, Gimenez-Ibanez S, García-Casado G, Porzel A, Pazos F, Boter M, Solano R. 2014. Rational design of a ligand-based antagonist of jasmonate perception. Nature Chemical Biology 10, 671–676. [DOI] [PubMed] [Google Scholar]
- Patkar RN, Benke PI, Qu Z, Chen YY, Yang F, Swarup S, Naqvi NI. 2015. A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nature Chemical Biology 11, 733–740. [DOI] [PubMed] [Google Scholar]
- Phillips AJL. 2002. Botryosphaeria species associated with diseases of grapevines in Portugal. Phytopathologia Mediterranea 41, 3–18. [Google Scholar]
- Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW. 2013. The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology 76, 51–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt WM, Huang R, Steel CC, Savocchia S. 2010. Identification, distribution and current taxonomy of Botryosphaeriaceae species associated with grapevine decline in New South Wales and South Australia. Australian Journal of Grape and Wine Research 16, 258–271. [Google Scholar]
- Reveglia P, Chini A, Mandoli A, Masi M, Cimmino A, Pescitelli G, Evidente A. 2018. Synthesis and mode of action studies of N-[(-)-jasmonyl]-S-tyrosin and ester seiridin jasmonate. Phytochemistry 147, 132–139. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual Review of Phytopathology 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. 2009. Enzymes in jasmonate biosynthesis – structure, function, regulation. Phytochemistry 70, 1532–1538. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. . 2010. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. 2004. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Turner WB, Aldridge DC. 1983. Fungal metabolites, vol. II. London: Academic Press. [Google Scholar]
- Úrbez-Torres JR. 2011. The status of Botryosphaeriaceae species infecting grapevine. Phytopathologia Mediterranea 50, 5–45. [Google Scholar]
- Van Niekerk JM, Fourie PH, Halleen F, Crous PW. 2006. Botryosphaeria spp. as grapevine trunk disease pathogens. Phytopathologia Mediterranea 45, 43–54. [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Feussner I. 2018. The oxylipin pathways: biochemistry and function. Annual Review in Plant Biology. In press. doi:10.1146/annurev-arplant-042817-040440. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Song S. 2017. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. Journal of Experimental Botany 68, 1303–1321. [DOI] [PubMed] [Google Scholar]
- Widemann E, Miesch L, Lugan R, Holder E, Heinrich C, Aubert Y, Miesch M, Pinot F, Heitz T. 2013. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. The Journal of Biological Chemistry 288, 31701–31714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam MG, Onkokesung N, Baldwin IT, Galis I. 2012. Jasmonoyl-L-isoleucine hydrolase 1 (JIH1) regulates jasmonoyl-L-isoleucine levels and attenuates plant defenses against herbivores. The Plant Journal 72, 758–767. [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. 1998. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. 2002. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. The Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, et al. . 2012. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences, USA 109, E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai QZ, Yan C, Li L, Xie DX, Li CY. 2017. Jasmonates. In: Li J, Li C, Smith SM, eds. Hormone metabolism and signaling in plants. Academic Press, 243–272. [Google Scholar]
- Zhang F, Yao J, Ke J, et al. . 2015. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Poudel AN, Jewell JB, Kitaoka N, Staswick P, Matsuura H, Koo AJ. 2016. Hormone crosstalk in wound stress response: wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. Journal of Experimental Botany 67, 2107–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.