Fig. 3.
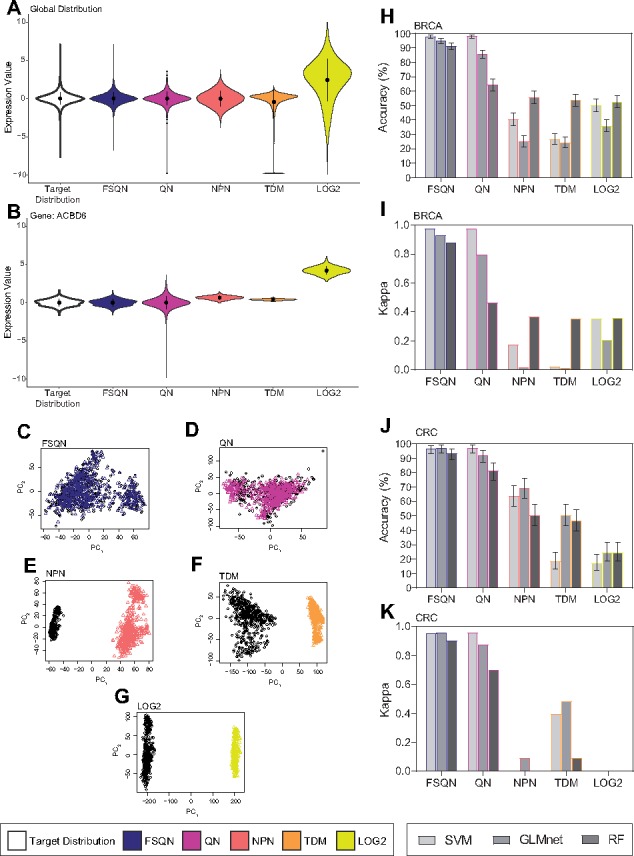
Normalization results with unscaled data. Different normalization techniques using unscaled BRCA data from both a global perspective of the data (A) and a local perspective from a randomly chosen gene (B). Principal components plot (PC1 v. PC2) for FSQN (C), MC (D), QN (E), NPN (F), TDM (G) and LOG2 (H) normalized BRCA RNA-seq and microarray data. Performance of classifications using normalized unscaled data classified by SVM, GLMnet and RF are reported as overall accuracy and kappa for the BRCA dataset (I, J) and the CRC dataset (K, L)
