Fig. 3.
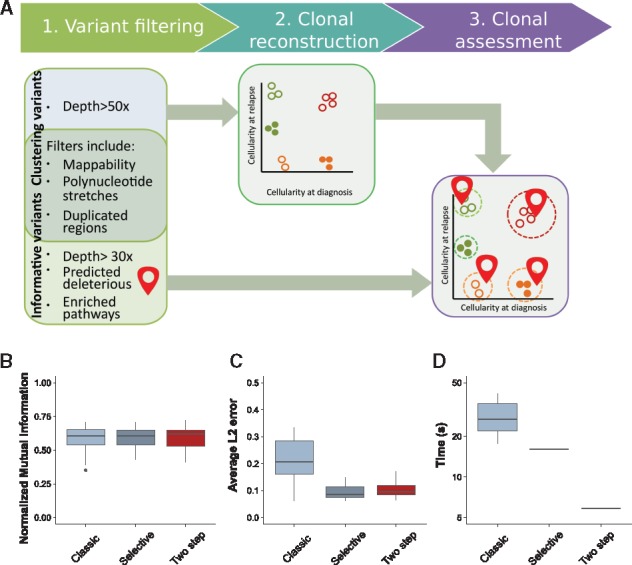
Assessment of the pipeline. (A) Overview of the general clonal reconstruction workflow: steps 1–3. (1) Variants are filtered to remove false positive calls; stringent filters are used to produce mutations that are further employed for clonal reconstruction (step 2), tolerant filters are used to detect functional mutations. (2) Variants that pass stringent filters and have genotype information assigned to the corresponding genomic loci are used as input to QuantumClone to reconstruct clonal populations. (3) Finally, possibly damaging mutations belonging to frequently altered pathways are mapped to the reconstructed clones. Quality of reconstruction. The pipeline aforementioned (two step), or a clustering using all variants called (classic) or a pipeline using only variants of biological interest and variants of high quality (selective) are assessed in terms of NMI (B), average L2 error (C) or computational time (D). The pipelines are evaluated on 20 simulations (Section 2)
