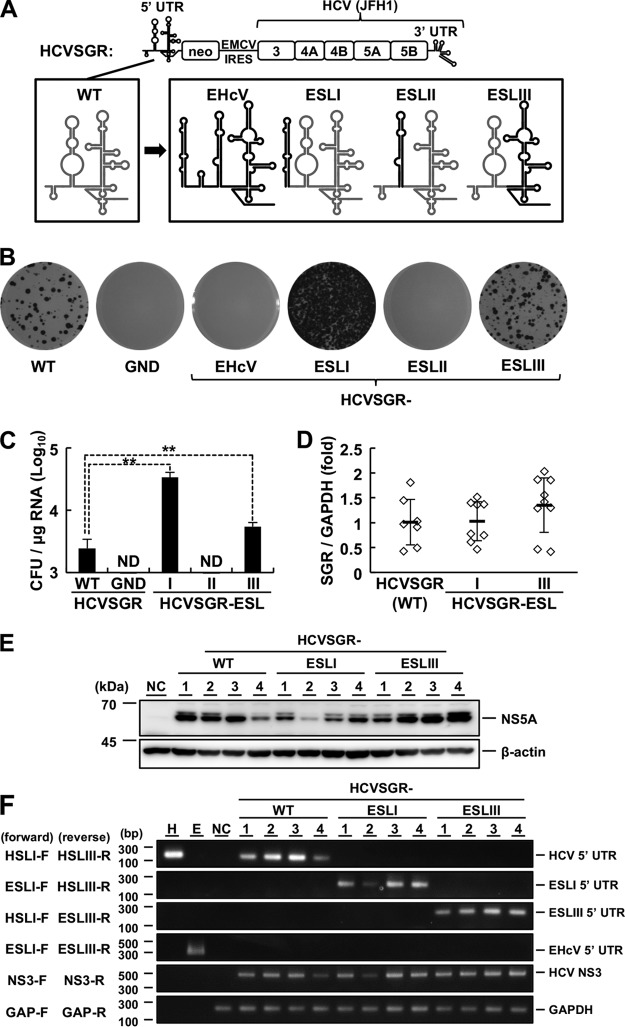
FIG 5.
Replication of the subgenomic replicon harboring the HCV-EHcV chimeric 5′ UTR. (A) Schematic diagram of HCVSGR and mutants. Domains I, II, and III-IV of the intrinsic HCVSGR 5′ UTR were replaced with domains I (ESLI), II (ESLII), and III (ESLIII), respectively, of the EHcV 5′ UTR. The domain structures of HCV and EHcV are indicated with gray and black lines, respectively. (B) The WT or mutant HCVSGR RNAs were subjected to a colony formation assay. The HCVSGR RNA carrying inactivated polymerase mutations (GND) was also subjected to the assay as a negative control. (C) Colony counts for the colony formation assay shown in panel B were estimated using ImageJ software to calculate CFU. ND, not detected. **, P < 0.01. (D) The amount of replicon RNA in each clone was estimated by qRT-PCR and normalized with the amount of GAPDH mRNA. The average value of each group is indicated as the mean (thick horizontal line) ± SD. (E) The cell lysates extracted from 4 clones in each group were subjected to Western blotting. Clone numbers are indicated at the top. NC indicates the lysate of nontransfected cells. (F) Total RNA extracted from the 4 clones in each group was subjected to RT-PCR analysis using the indicated primer pairs (Table 1). Clone numbers are indicated at the top. In vitro-transcribed RNAs including the HCV 5′ UTR (lane H) or EHcV 5′ UTR (lane E) were used as positive controls. The total RNA extracted from the nontransfected cells (NC) was used as a negative control. The data are representative of the results of three independent experiments. The error bars indicate standard deviations.