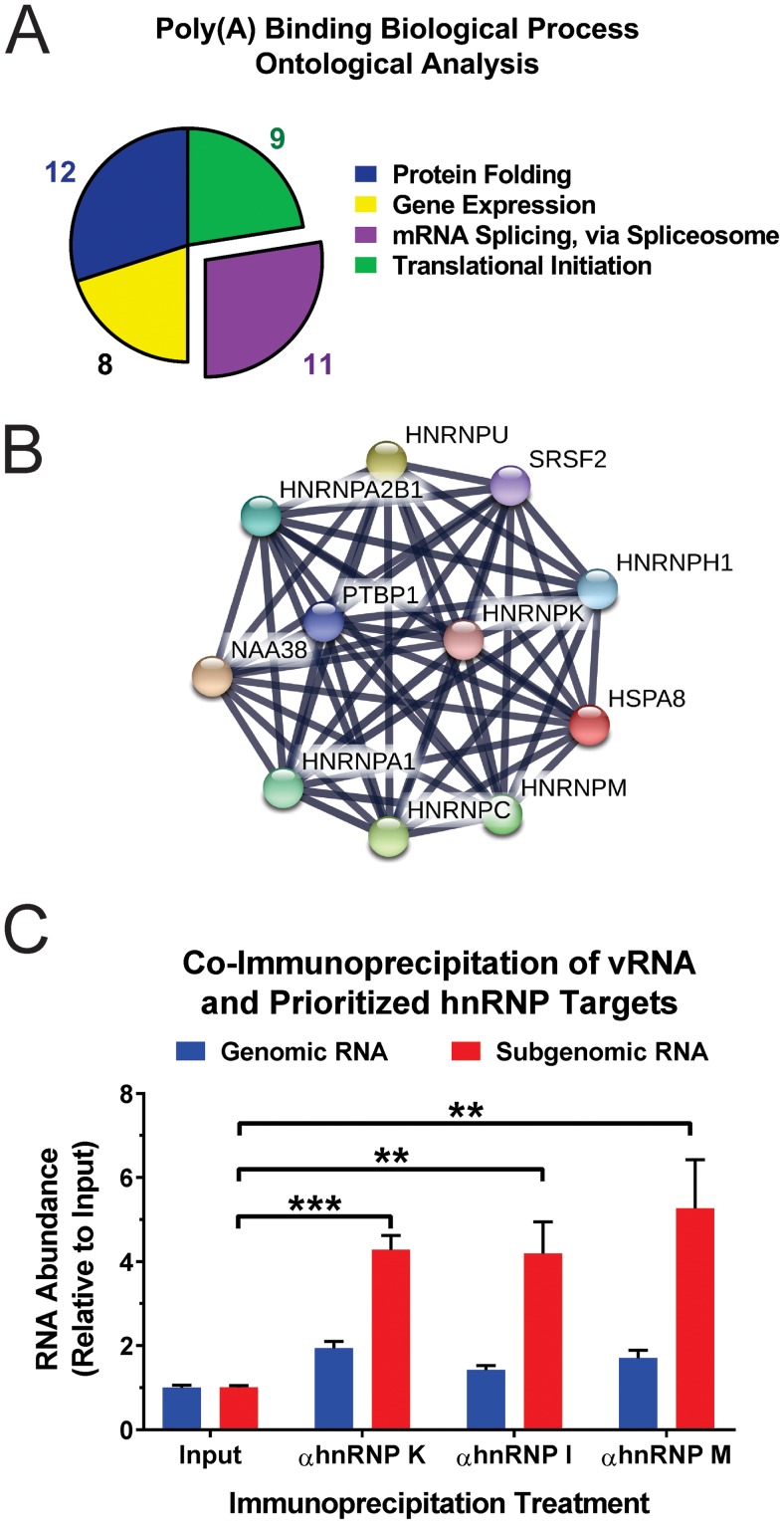
FIG 2.
Ontological and interaction analysis of SINV CLAMP poly(A) RNA-binding proteins. (A) The SINV CLAMP proteins identified by DAVID as belonging to the poly(A) RNA-binding molecular function group were assessed to identify enriched biological processes. Shown is a pie graph of the 4 biological process ontological groups statistically enriched as determined by DAVID analyses. (B) Interaction map of the SINV CLAMP-identified interactants grouped into the mRNA splicing via spliceosome biological process ontological group. (C) Immunoprecipitation of cross-linked SINV vRNP complexes derived from 293HEK cells infected with wild-type SINV indicated that the prioritized hnRNP-vRNA interactants are direct interactions. Quantitative detection of the enrichment of the individual vRNA species relative to input levels was accomplished using qRT-PCR. The values shown are the means of at least three biological replicates, with the error bars representing standard deviations of the mean. Statistical significance was determined by Student's t test; **, P < 0.01; ***, P < 0.001.