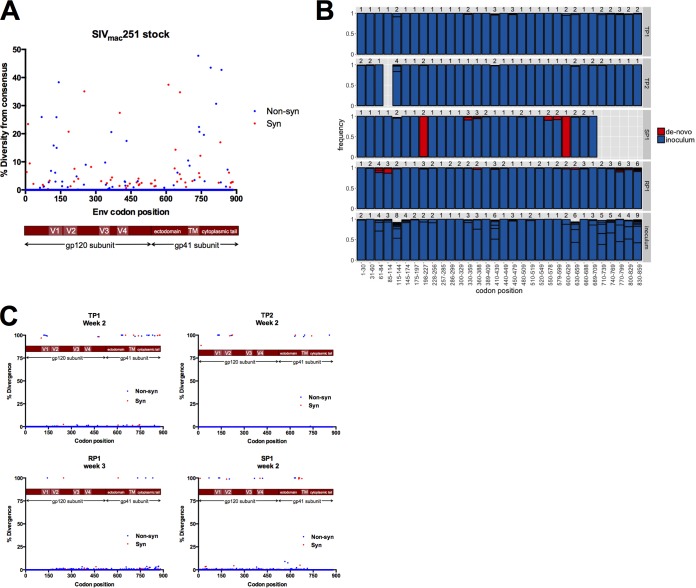
FIG 2.
Nonsynonymous and synonymous diversity in the uncloned SIVmac251 stock followed by genetic bottleneck at transmission. (A) The level of nonsynonymous (blue) and synonymous (red) diversity was determined for every codon in Env in the uncloned SIVmac251 stock inoculum. Nonsynonymous diversity was determined as the frequency of codons with a nonsynonymous change relative to the majority codon of the stock consensus sequence, and synonymous diversity was determined as the frequency of codons with a synonymous change relative to the majority codon of the stock consensus sequence. (B) We split Env into 90-nucleotide windows (30 codons). For each animal and the stock inoculum, we collected NGS reads spanning each window and characterized the number of variants. For the animals, sequences from the first sample time postinfection were considered. Each bar shows particular variants and their frequency (boxed in black). Variants either were found in the inoculum (blue) or not (red). Variants not found in the inoculum, labeled de novo, could either have been generated during infection through mutation or represent low-frequency inoculum variants that were not identified. Missing bars represent windows with poor NGS coverage, with <100 high-quality reads. Bars that do not reach a frequency of 1 reflect variants with a frequency <1%, which are not shown for readability. (C) Divergence was determined as the frequency of a majority codon (frequency of 50% or more) at the sampled time point that differed from the majority codon of the SIVmac251 stock inoculum. Nonsynonymous sites are reported in blue, and synonymous sites are reported in red.