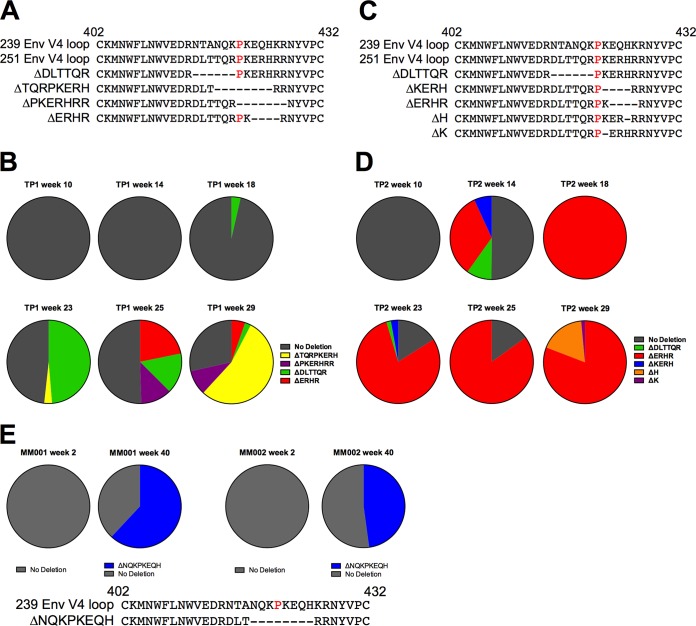
FIG 5.
Evolution of in-frame deletions in SIV Env V4 loop observed in TP1 and TP2. (A to D) Deletions in V4 and frequencies for animal TP1 (A and B) and animal TP2 (C and D). (A and C) Deletions are depicted relative to the V4 amino acid sequence of SIVmac239 and the consensus of the SIVmac251 stock inoculum. In-frame multiple-amino-acid deletions flank or overlap the P421 site (highlighted in red in the 239 and 251 V4 reference sequences). (B and D) The relative frequency of deletion variants detected in the reads spanning the entire V4 loop are reported at 10, 14, 18, 23, 25, and 29 weeks p.i. Reads lacking in-frame deletions were reported as having no deletions (in gray). (E) Viral RNA from MM001 and MM002 were deep sequenced as part of a separate study (A. K. Hill, S. Ita, R. Newman, and W. E. Johnson, unpublished data). The relative frequency of deletion variants detected in the reads spanning the V4 loop from amino acid positions 410 to 439 at 2 and 40 weeks p.i. are reported. Reads that did not possess any in-frame deletions were reported as no deletion (in gray).