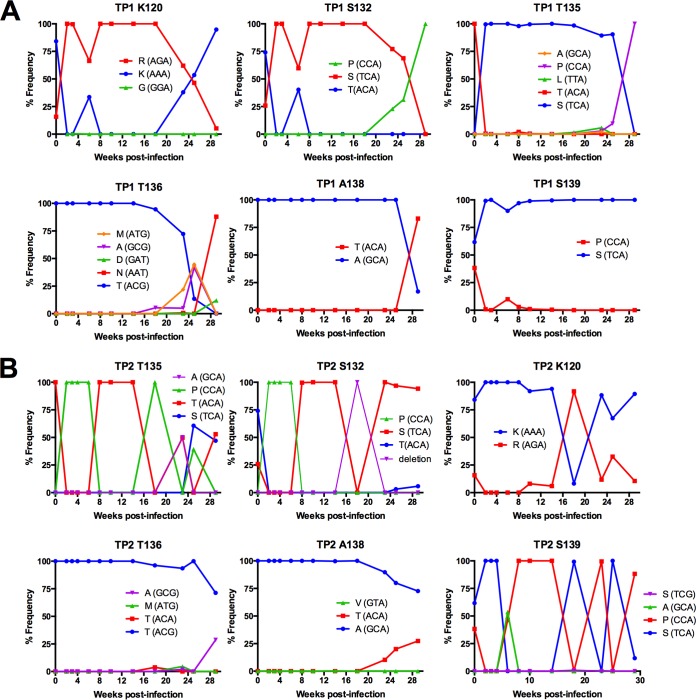
FIG 7.
Evolution of amino acid substitutions in SIV Env V1 loop in TP1 and TP2. The frequency of codons was determined from deep sequence reads of each sample assembled to the de novo consensus sequence of the same sample using the V-Profiler software program (Broad Research Institute). The six sites within the hypervariable region of V1 are plotted over the first 29 weeks of in vivo viral replication and are representative of sites with the highest codon variation for TP1 (A) and TP2 (B).