ABSTRACT
HIV-1 downregulates human leukocyte antigen A (HLA-A) and HLA-B from the surface of infected cells primarily to evade CD8 T cell recognition. HLA-C was thought to remain on the cell surface and bind inhibitory killer immunoglobulin-like receptors, preventing natural killer (NK) cell-mediated suppression. However, a recent study found HIV-1 primary viruses have the capacity to downregulate HLA-C. The goal of this study was to assess the heterogeneity of HLA-A, HLA-B, and HLA-C downregulation among full-length primary viruses from six chronically infected and six newly infected individuals from transmission pairs and to determine whether transmitted/founder variants exhibit common HLA class I downregulation characteristics. We measured HLA-A, HLA-B, HLA-C, and total HLA class I downregulation by flow cytometry of primary CD4 T cells infected with 40 infectious molecular clones. Primary viruses mediated a range of HLA class I downregulation capacities (1.3- to 6.1-fold) which could differ significantly between transmission pairs. Downregulation of HLA-C surface expression on infected cells correlated with susceptibility to in vitro NK cell suppression of virus release. Despite this, transmitted/founder variants did not share a downregulation signature and instead were more similar to the quasispecies of matched donor partners. These data indicate that a range of viral abilities to downregulate HLA-A, HLA-B, and HLA-C exist within and between individuals that can have functional consequences on immune recognition.
IMPORTANCE Subtype C HIV-1 is the predominant subtype involved in heterosexual transmission in sub-Saharan Africa. Authentic subtype C viruses that contain natural sequence variations throughout the genome often are not used in experimental systems due to technical constraints and sample availability. In this study, authentic full-length subtype C viruses, including transmitted/founder viruses, were examined for the ability to disrupt surface expression of HLA class I molecules, which are central to both adaptive and innate immune responses to viral infections. We found that the HLA class I downregulation capacity of primary viruses varied, and HLA-C downregulation capacity impacted viral suppression by natural killer cells. Transmitted viruses were not distinct in the capacity for HLA class I downregulation or natural killer cell evasion. These results enrich our understanding of the phenotypic variation existing among natural HIV-1 viruses and how that might impact the ability of the immune system to recognize infected cells in acute and chronic infection.
KEYWORDS: transmitted/founder virus, HIV-1, Nef, Vpu, quasispecies, human leukocyte antigen class I, HLA, NK cells, human immunodeficiency virus, natural killer cells
INTRODUCTION
Human leukocyte antigen (HLA) class I molecules loaded with peptides are downregulated from the surface of human immunodeficiency virus type 1 (HIV-1)-infected CD4 T cells (1). Nef, an HIV-1-encoded accessory protein, mediates downregulation of HLA-A and HLA-B (2, 3), thus protecting against HIV-specific CD8 T cell recognition (4). HLA-C was previously thought to be left on the surface of infected cells in order to inhibit natural killer (NK) cell-mediated viral suppression through binding of killer cell immunoglobulin-like receptors (KIR) as inhibitory ligands (3, 5–8). However, a recent discovery changed this paradigm by demonstrating that Vpu, another HIV-1 accessory protein with functions that overlap those of Nef, downregulates HLA-C, a finding previously obscured by wide usage of laboratory-adapted strains lacking such an ability (9). This highlights the importance of studying patient-derived uncultured viruses when possible.
HLA class I expression can impact NK and CD8 T cell recognition of virally infected cells, thus modulating adaptive and innate immunity. HLA-A and HLA-B present viral peptides primarily to CD8 T cells by interacting with their T cell receptors. Reducing surface expression of HLA-A and HLA-B prevents epitope-specific recognition of infected cells by CD8 T cells which could otherwise suppress virus replication (4). Strong experimental evidence supporting this phenomenon is demonstrated by the in vitro selection of viruses with efficient Nef-mediated HLA-A downregulation capacity when passaged in the presence of Gag-specific CD8 T cell clones (10). The in vivo relevance has been examined in experiments of simian immunodeficiency virus (SIV)-infected rhesus macaques. SIVMAC239 nef mutants deficient in major histocompatibility complex class I (MHC-I) downregulation revert early in infection (11), and SIVMAC239-infected rapid progressors exhibited a 2-fold higher level of MHC-I downregulation on infected cells ex vivo, which was associated with lower peptide-specific T cell responses (12). In contrast, a recent study found little MHC-I downregulation on SIVMAC239-infected cells ex vivo using a pan-MHC-I antibody (13). Peptide-specific NK KIR interactions with HLA-A and HLA-B alleles also impact HIV infection (14, 15) and are associated with variations in viral control and disease progression (16–19). Although the consequences of HLA-C expression are less well defined, both NK and CD8 T cell responses are impacted by HLA-C. Higher HLA-C expression in infected individuals correlates with slower CD4 T cell decline, increased CD8 T cell responses, and selection of HLA-C-associated viral escape mutations (20). NK KIR interactions with HLA-C can also drive HIV sequence-based adaptations (21).
The downregulation of HLA class I molecules from the surface of infected cells may affect the establishment of infection. Both HLA class I allele sharing between heterosexual transmission pairs and HLA class I homozygosity in mother-child pairs increased the risk of infection, indicating a role for HLA class I molecules during transmission (22, 23). Combinations of HLA-A, HLA-B, and HLA-C alleles with specific NK KIR alleles have been associated with protection from HIV acquisition (24–28), and NK cells have also been implicated in the SIV macaque model, where elevated CD56+ NK cell frequencies were associated with relative protection from SIVMAC251 challenge when interferon alpha was preadministered (29). Thus, viral characteristics that modulate HLA class I expression may play a role in HIV acquisition and dissemination.
Acquisition of HIV-1 via the heterosexual route is characterized by a low chance of infection per mucosal exposure (30, 31), accompanied by a genetic bottleneck that leads to one or a few viral variants establishing infection in a new individual (31, 32). Understanding the forces that determine which viruses break through has implications for prevention strategies, including vaccines. Although chance certainly influences which viral variants become transmitted/founder (TF) viruses, several studies have reported signature characteristics of transmitted/founder viruses (31–37). A study of 137 subtype C-infected epidemiologically linked Zambian transmission pairs found that Gag, Pol, and Nef consensus sequences are preferentially transmitted, a signature emphasized in Nef functional domains associated with HLA class I downregulation, suggesting in vivo viral fitness plays a role in the transmission bottleneck (38). The genetic selection for consensus amino acids was confirmed across the entire genome sequence from six subtype C-infected transmission pairs; however, virus-mediated HLA class I downregulation was not examined in that study (35).
Prior studies have not observed differences in HLA class I downregulation by subtype C viruses from acute and chronic infection (9, 39, 40). However, viruses derived from heterosexual transmission pairs with the source quasispecies as a comparison were not available. Moreover, the three major HLA class I molecules, HLA-A, HLA-B, and HLA-C, were not addressed together in primary cells. Most prior studies characterized HLA class I downregulation in cell lines with Nef expression vectors and chimeric viruses, which may alter physiologic expression levels, or exclude the effect of coevolved Vpu proteins and other areas of the genome, influencing protein expression generally (39, 41–47). Additionally, the extent to which there is within-host functional diversity of HLA class I downregulation, which may reflect selection by the cellular immune system, remains to be established (10, 42, 47, 48).
In the study described here, we examined HLA-A, HLA-B, and HLA-C downregulation using 53 HIV-1 variants: 40 infectious molecular clones previously constructed from plasma virus sequences from 12 individuals in six subtype C heterosexual transmission pairs, 12 subtype C Gag-MJ4 chimeras varying in replicative capacities (RC), and a standard laboratory subtype B strain, NL4-3 (35, 49, 50). We also investigated the relationship between HLA expression and the ability to evade NK cell suppression in vitro. We found a range of HLA class I downregulation capacities (1.3- to 6.1-fold) of viruses from within and between individuals. The ability to downregulate HLA class I molecules, including HLA-C, was similar between source quasispecies and transmitted/founder viruses. Although replicative capacity influenced evasion of NK-mediated suppression, HLA-C downregulation was significantly associated with the susceptibility of variants to NK cell suppression. These data could be important to consider in cure and vaccine strategies aiming to induce CD8 and NK cell responses.
RESULTS
Variation of Nef and Vpu proteins of subtype C viruses from transmission pairs.
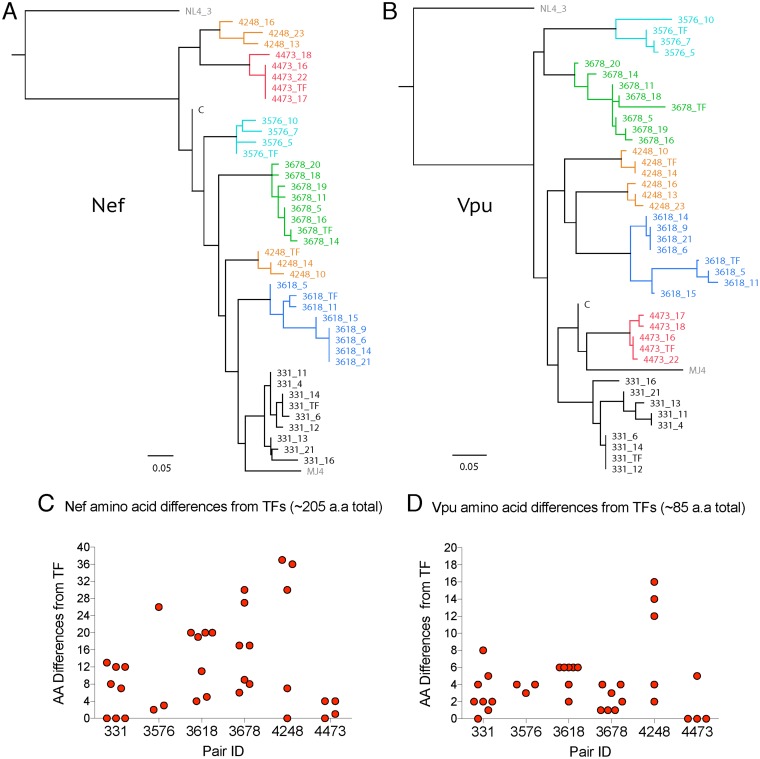
We previously sequenced and cloned 40 full-length infectious molecular clones from six subtype C transmission pairs from the Zambia-Emory HIV Research Project (35, 51). To display the diversity of Nef and Vpu proteins in these viruses, we constructed phylogenetic trees of Nef and Vpu amino acid sequences, along with the laboratory strains NL4-3 and MJ4 (Fig. 1A and B). Viruses from within donor partners harbored a mean of 10 amino acid differences in Nef (range, 0 to 35), whereas viruses from different donor partners had a mean of 37 Nef amino acid differences (range, 21 to 59). Pair 4248 had two distinct Nef branches due to deletions and insertions in the N-terminal anchor domain (for amino acid alignments of Nef and Vpu sequences, see Data Set S1 in the supplemental material). For Vpu, a mean of 5 amino acid differences was observed in viruses from within an infected individual (range, 0 to 17), while between individuals a mean difference of 19 amino acids was observed (range, 10 to 26). Nontransmitted (NT) variants differed to various degrees from their cognate transmitted/founder virus in each pair in their Nef and Vpu amino acid sequences (Fig. 1C and D). The amino acid diversity found throughout Nef and Vpu between related and unrelated subtype C primary variants highlighted the potential for heterogeneity of HLA class I downregulation activities within a host quasispecies, between hosts infected with viruses of the same subtype, and across the transmission bottleneck.
FIG 1.
Diversity of Nef and Vpu in characterized viruses. Viral variants from donors of transmission pairs are labeled as pair ID followed by variant number, while recipient transmitted/founder viruses are labeled TF. Each pair is set off by color. The laboratory strains are in gray, while the subtype C LANL consensus is labeled C. BioNJ tree of phylogenetic relationships of Nef (A) and Vpu (B) amino acid (aa) sequences of transmitted/founder and nontransmitted donor variants (NT), along with one subtype C (MJ4) and one subtype B (NL4-3) laboratory strain. The LANL subtype C consensus is included for reference. Number of amino acid differences from the transmitted/founder for each NT donor variant from each transmission pair for Nef (C) and Vpu (D).
HLA class I downregulation of subtype C variants from transmission pairs and laboratory strains.
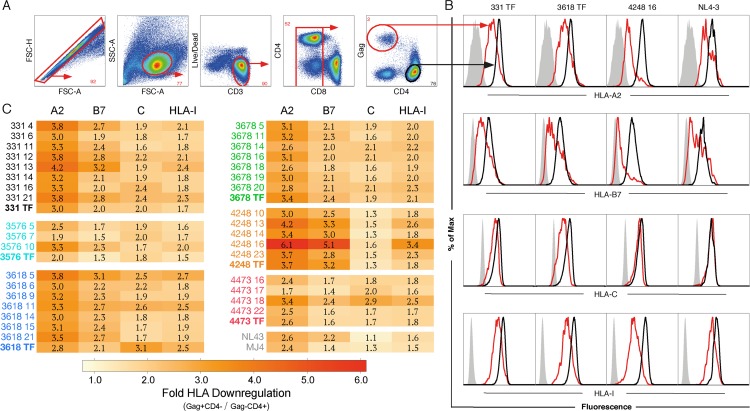
The extent to which authentic primary subtype C viruses differ in the capacity to downregulate HLA-A, HLA-B, and HLA-C in primary cells, including those from transmission pairs, is unclear. To measure HLA-A, -B, and -C downregulation simultaneously on primary cells by flow cytometry, we used commercially available serotype-specific antibodies for cells coexpressing HLA-A2 and HLA-B7, added an HLA-C-specific antibody (DT9) to measure HLA-C expression, and included a pan-HLA class I antibody (W6/32) to measure total class I expression. For the majority of the in vitro experiments described here, we utilized leukapheresed cells from an individual expressing both HLA-A2 and HLA-B7 alleles (HLA typing of HIV-negative individual number 3 confirmed the presence of both alleles; Data Set S2). Primary cells were infected in vitro and stained at day seven, which enabled collection of data for both low- and high-replicative-capacity variants. Gag+ CD4− T cells were gated and compared to Gag− CD4+ T cells, since HLA class I downregulation could be robustly detected when CD4 was maximally downregulated and Gag expression was peaking (Fig. 2A), as has been described elsewhere (52). Consistent with this, Gaglow CD4+ cells overlapped in expression of HLA class I with Gag− CD4+ cells and thus were not shown as a separate population. The median fluorescent intensity (MFI) and percent HLA-negative cells correlated (P < 0.0001) for HLA-A (r = 0.78), HLA-B (r = 0.93), HLA-C (r = 0.91), and total HLA-I (r = 0.92); thus, MFI was used to measure downregulation in subsequent analyses.
FIG 2.
HLA downregulation of infected cells in vitro. Infectious molecular clones from six transmission pairs along with two laboratory strains (MJ4, subtype C; NL4-3, subtype B) were assayed for the ability to downregulate HLA-I by flow cytometry with antibodies recognizing HLA-A2, HLA-B7, HLA-C, and pan-HLA-I. The gating strategy is shown as an example from 331 TF (A), and histograms showing CD4− Gag+ (red), CD4+ Gag− (black), and HLA negative controls (gray) are shown for 4 variants that show a range of activities for HLA-A2 (top row), HLA-B7 (row 2), HLA-C (row 3), and HLA-I (bottom row) downregulation (B). (C) Mean values from pooled triplicates averaged from 2 to 5 experiments of the fold downregulation of HLA by MFI on infected T cells, as described for panels A and B, compared to uninfected CD4+ T cells. Standard errors for each HLA class I factor are provided in Materials and Methods. Each viral clone is denoted with a pair ID followed by variant number when amplified from the donor partner or TF for variants derived from the recipient. HLA-A2, A2; HLA-B7, B7; HLA-C, C; pan-HLA-I, HLA-I; TF, transmitted/founder; NT, nontransmitted donor variant.
The extent of downregulation of HLA-A, HLA-B, HLA-C, and total HLA-I on infected cells is illustrated in Fig. 2B, where representative histogram plots from four variants display a range of viral phenotypes (Fig. 2B). The subtype B laboratory strain NL4-3 did not measurably downregulate HLA-C, as described previously (9), while the transmitted/founder from pair 3618 displayed the highest level of HLA-C downregulation of the variants tested (Fig. 2B). Variant 4248 TF 16 showed relatively low levels of HLA-C downregulation yet robust HLA-A and -B downregulation, consistent with discrete functions of Nef and Vpu (Fig. 2B).
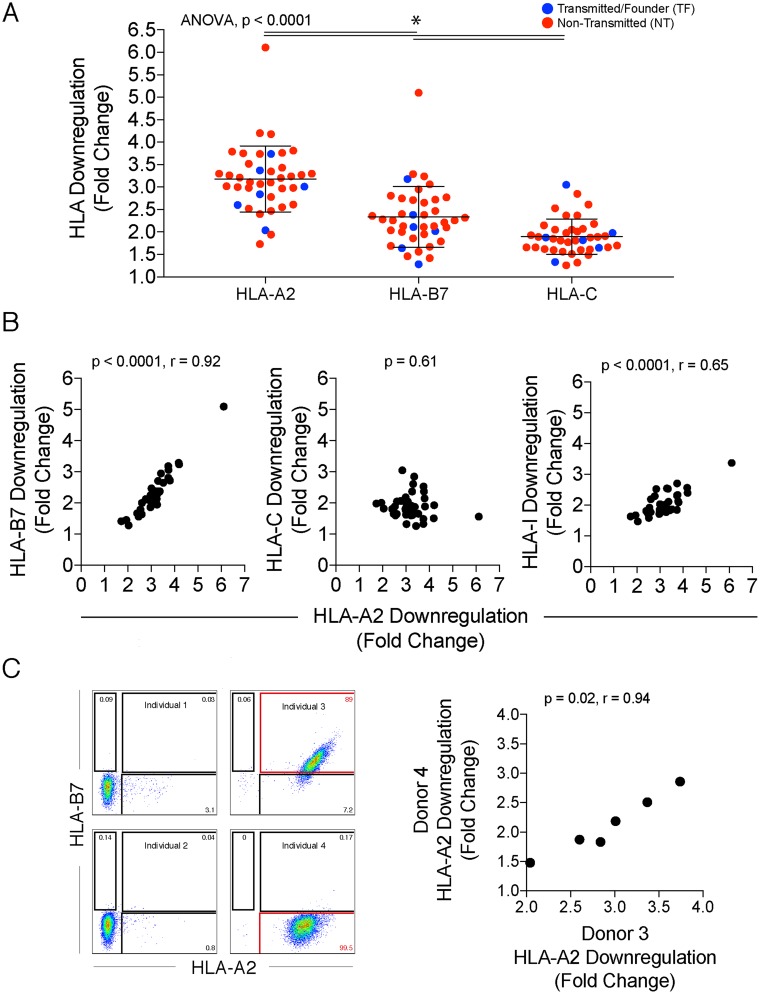
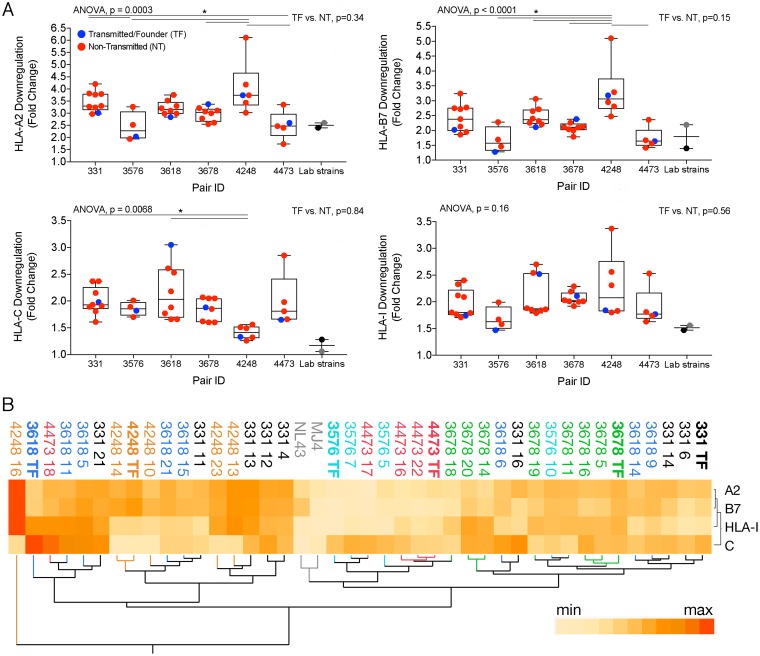
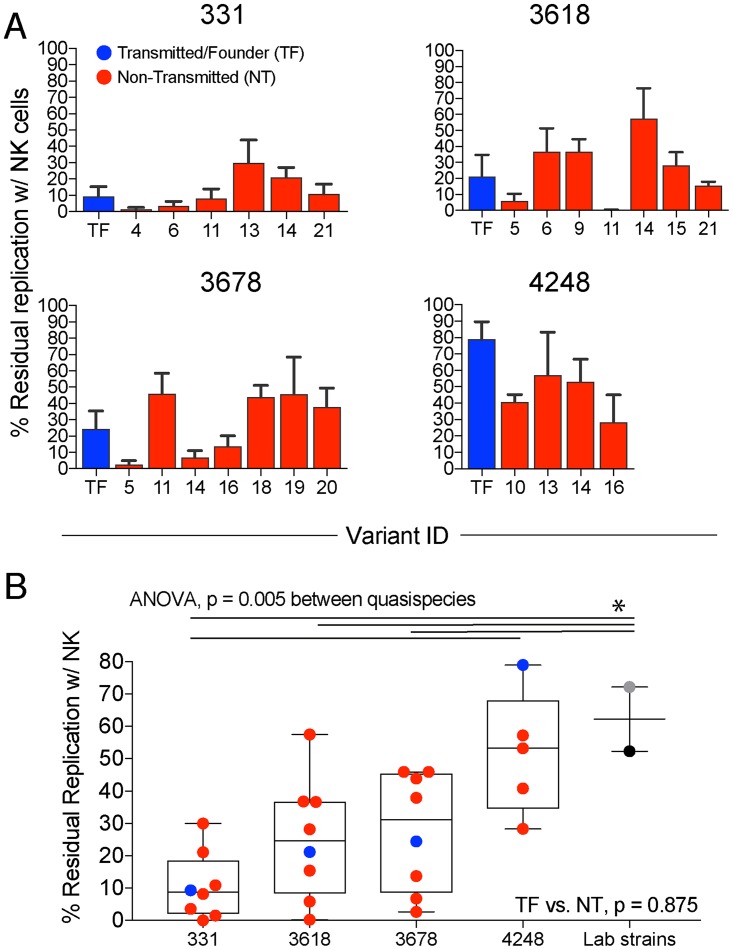
The mean fold downregulation from multiple replicates and experiments is shown in a heat map in Fig. 2C for HLA-A, HLA-B, HLA-C, and total HLA-I (Fig. 2C). HLA-A2 was downregulated by the largest fold change in expression on infected cells by MFI compared to that on CD4+ Gag− T cells, with a range of 1.7 to 6.1 (median, 3.2) (Fig. 3A). HLA-C had a more limited range of downregulation by fold change (range, 1.3 to 3.1; median, 1.9), potentially due to the lower level of expression of HLA-C on uninfected primary cells (Fig. 2B and 3A) (6). Downregulation of HLA-B7, which ranged from 1.3- to 5.1-fold (median, 2.3), was highly correlated with that of HLA-A2 (r = 0.92, P < 0.0001) (Fig. 3A and B). In contrast, there was no correlation between HLA-A2 and HLA-C downregulation (P = 0.61) (Fig. 3B), consistent with these molecules being downregulated by different viral gene products. Peripheral blood mononuclear cells (PBMC) from a different uninfected HLA-A2+/HLA-B7− individual (sequence confirmed in Data Set S2) yielded similar results for the six transmitted/founder variants analyzed (Fig. 3C). The slight increase in fold downregulation for the PBMC of individual 3 versus 4 could be because individual 3 is homozygous at HLA-A (Data Set S2). Transmitted/founder variants generally, but not universally, exhibited lower downregulation for HLA-A (P = 0.34) and HLA-B (P = 0.15), and the difference was not significant (Fig. 4A). Significant differences between quasispecies from different infected individuals were observed for HLA-A (P = 0.0003), HLA-B (P < 0.0001), and HLA-C (P = 0.007) downregulation (Fig. 4A), demonstrating divergence of HLA downregulation capacity between quasispecies. NL4-3 and MJ4 exhibited low levels of HLA-I downregulation (Fig. 4A).
FIG 3.
Relationship of HLA class I molecules on infected cells. (A) Overall HLA class I downregulation for the tested primary variants showing the means and standard deviations. Bars with asterisks above mark significant differences. TF are blue and NT are red. (B) Spearman correlation analysis between HLA-A2 and HLA-B7 downregulation (left), HLA-A2 and HLA-C downregulation (middle), and HLA-A2 and total HLA-I (right). (C, left) Identification of HLA-A2+ HLA-B7+ and HLA-A2+ HLAB7− cells by flow cytometry by screening of PBMC from 4 different individuals as examples. (Right) Spearman correlation of HLA-A2 downregulation in two different PBMC donors averaged from multiple experiments using TF variants.
FIG 4.
Quasispecies-level HLA class I downregulation and clustering by phenotype. (A) Box-and-whisker plots showing median and minimum to maximum per transmission pair (x axis) for downregulation of HLA-A2 (top left), HLA-B7 (top right), HLA-C (bottom left), and total HLA-I (bottom right). Transmitted viruses are in blue, nontransmitted variants are in red, NL4-3 is in gray, and MJ4 is in black. Lines and asterisks represent statistical significance at a P value of <0.05, and all y axes depict fold change of CD4− Gag+ MFI divided by CD4+ Gag− MFI. (B) Hierarchical clustering analysis of variants by HLA downregulation level independent of their genetic relatedness. Each variant is labeled and colored per transmission pair as described for Fig. 2C.
Since viruses from different infected individuals often displayed similar downregulation capacities and related variants appeared by visual inspection of the heat map to have similar phenotypes (Fig. 2C), we performed hierarchical clustering to determine phenotypic linkages between variants using only their HLA functional footprint (Fig. 4B). Although transmitted/founder variants did not cluster or have a common signature, related variants often, but not always, grouped together. For instance, 4248 TF 10 and 14, which appear on the same cluster by their Nef and Vpu sequences on the phylogenetic tree (Fig. 1A and B), also clustered by their HLA class I downregulation phenotypic signatures (Fig. 4B). Likewise, pair 331 TF 6 and 14 have the most similar Nef and Vpu sequences from that transmission pair and are clustered by their HLA phenotype despite having an almost 20-fold range of infectivities (331 TF, 3.5 × 10−3; 331 TF 6, 4.3 × 10−4; 331 TF 14, 2.5 × 10−3) and a more than 10-fold range of replicative capacities (331 TF, 1.12; 331 TF 6, 0.15; 331 TF 14, 1.58), which were previously determined for these viruses (35). Moreover, the laboratory strains NL4-3 and MJ4 clustered together with a pattern of low downregulation despite their differences in sequence, subtype, and coreceptor usage, which might reflect laboratory adaptation. Both convergence and divergence of phenotypes between quasispecies is evident from the interspersion of some variants throughout the clusters, as was the case for 331 TF 11, for instance, which clustered next to 3618 TF 15 (Fig. 4B). In 2/6 cases, groups of viruses from the same quasispecies were in two distant clusters (331 and 3618) (Fig. 4B). However, in 4/6 cases, >75% of variants from a given quasispecies fell into just one of four major clusters, demonstrating the general relationship between genetic and phenotypic characteristics as they relate to HLA class I downregulation (Fig. 4B).
Genotypic correlates of HLA class I downregulation.
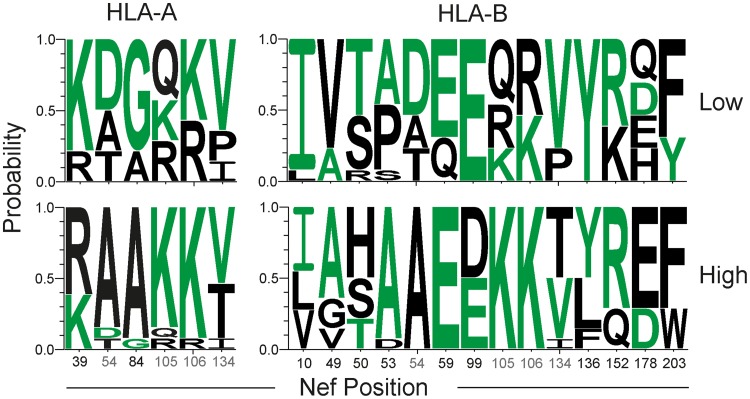
To identify amino acid positions potentially contributing to differential HLA class I downregulation, the highest- and lowest-performing terciles in HLA-A, HLA-B, HLA-C, and total HLA class I downregulation were compared for amino acid signatures by Sequence Harmony analysis (53). Significant positions, identified as described in Materials and Methods, were found throughout Nef (Table 1), although none were identified for HLA-C-associated Vpu sites. A number of amino acids identified here were previously reported to be involved in HLA class I downregulation, including Nef positions 84 (10), 99 (41), 106 (39), 134 (39), 136 (10), and 203 (7, 10, 41). Sites matching the consensus sequence were not always associated with an increased capacity to downregulate HLA class I when grouped by HLA-A or HLA-B downregulation (Fig. 5). Furthermore, in contrast to the impact on downregulation observed following early escape from CTL responses (42), HLA-I polymorphisms linked statistically to donor or recipient partner HLA haplotypes were not specifically associated with viruses with reduced Nef-mediated HLA class I downregulation capacity, and no correlation was observed between the distance to the subtype C consensus in Nef and HLA class I downregulation (data not shown).
TABLE 1.
Amino acids of primary viruses significantly associated with HLA class I downregulation by Sequence Harmony analysis
| Nef positiona | Vpu positiona | SH scoreb | Z scorec | Groupingd |
|---|---|---|---|---|
| 10 | 0.67 | −3.6 | HLA-B | |
| 39 | 0.88 | −3.46 | HLA-A | |
| 49 | 0.65 | −5.21 | HLA-B | |
| 50 | 0.65 | −4.78 | HLA-B | |
| 53 | 0.64 | −6.42 | HLA-B | |
| 54 | 0.75 | −3.52 | HLA-A | |
| 54 | 0.43 | −9.04 | HLA-B | |
| 59 | 0.83 | −3.36 | HLA-B | |
| 84 | 0.58 | −9.05 | HLA-A | |
| 99 | 0.69 | −5.7 | HLA-B | |
| 99 | 0.87 | −3.18 | HLA-I | |
| 105 | 0.75 | −4.11 | HLA-A | |
| 105 | 0.43 | −8.51 | HLA-B | |
| 106 | 0.87 | −3.17 | HLA-A | |
| 106 | 0.66 | −6.45 | HLA-B | |
| 134 | 0.69 | −3.1 | HLA-A | |
| 134 | 0.56 | −5.68 | HLA-B | |
| 134 | 0.7 | −3.26 | HLA-I | |
| 136 | 0.69 | −3.73 | HLA-B | |
| 152 | 0.66 | −4.06 | HLA-B | |
| 178 | 0.64 | −3.09 | HLA-B | |
| 203 | 0.7 | −3.56 | HLA-B | |
| 15 | 0.85 | −3.99 | HLA-I | |
| 18 | 0.82 | −4.06 | HLA-I | |
| 77 | 0.83 | −4.13 | HLA-I |
Position numbers based on the alignment in Data Set S1 in the supplemental material.
Sequence Harmony (SH) score. Lower values equate to greater amino acid segregation.
Standard deviations below the mean of 100 randomizations of sequences. More negative values are more significant.
HLA class I downregulation values from Fig. 2C were divided into terciles 1 (low) and 3 (high) for each shown measured factor.
FIG 5.
Amino acids of viruses with high and low levels of HLA-A and HLA-B downregulation. Shown is a Weblogos display of the Nef amino acid sequence composition of primary viruses at the lowest and highest terciles for HLA-A and HLA-B downregulation. Sequence Harmony analysis identified the shown positions with a significant Z score, determined as described in Materials and Methods. A full listing of amino acids is presented in Table 1. Green amino acids denote the subtype C consensus amino acid at that position, and gray numbers are Nef positions common to virus groupings by HLA-A and HLA-B. Position numbers are based on the alignment in Data Set S1 in the supplemental material.
Natural killer cell suppression of subtype C variants and Gag-MJ4 chimeras in vitro.
NK cells, as a component of innate immunity, act at the portals of entry and during the initial phases of viral dissemination. Whether the capacity of primary HIV-1 variants to downregulate HLA-C, which can act as an inhibitory ligand for NK cells, influences NK cell susceptibility in primary infected cells has not been demonstrated. We therefore examined the ability of different variants to evade NK cell suppression in vitro within and between quasispecies, as well as across transmission pairs.
To investigate the relationship between NK-mediated suppression of virus replication and HLA-C downregulation, we modified a previously published assay assessing NK-mediated viral suppression (54) by comparing viral strains rather than NK cell donors. The assay employs primary CD4+ T cells infected in vitro to serve as targets, while autologous CD56+ NK cells isolated in parallel serve as effectors. We examined a representative subset of 29 variants that included matched transmitted/founder and nontransmitted variants, as well as NL4-3, MJ4, and Gag-MJ4 chimeras, with a range of HLA downregulation phenotypes and replicative capacities.
A range of levels of NK cell-mediated suppression of primary virus replication was observed at 7 days postinfection using a multiplicity of infection equal to 1.0, with between 1.5 and 79% residual replication (median, 28%) (Fig. 6A) in the presence of NK cells, equivalent to an effector-to-target (E:T) ratio of 1:10. Laboratory strains NL4-3 and MJ4 were far less susceptible to NK suppression than primary viruses from different quasispecies (Fig. 6B). Although there was overlap between viruses from different individuals, a significant difference was observed between pairs 331 and 4248 (Fig. 6B).
FIG 6.
NK cell suppression of virus growth in vitro. (A) TF (blue) and NT (red) residual replication in the presence of NK cells following infection at a multiplicity of infection of 1.0 is shown for four pairs. Each variant is listed on the x axis, and the y axis is calculated as the percentage of replication in the presence of NK cells compared to the no-NK controls equivalent to an effector-to-target ratio of 1:10, as further described in Materials and Methods. The error bars represent the standard errors of the means. (B) Mean quasispecies NK susceptibility, including that for laboratory strains NL4-3 (gray) and MJ4 (black). Lines and asterisks represent a P value of <0.05 using ANOVA followed by Tukey's multiple-comparison test. Wilcoxon test was done to compare TF and NT median for the pairs.
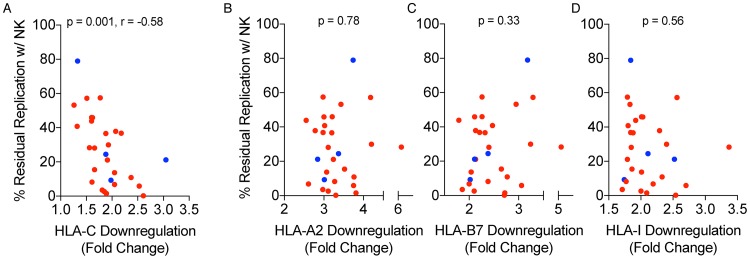
In primary viruses, the level of HLA-C downregulation significantly correlated with the level of NK suppression of virus release (P = 0.001, r = −0.58) (Fig. 7A), and this association held when testing a different subset of variants in PBMC from a different uninfected person (P = 0.059, r = −0.57). In contrast, no association was observed with downregulation of HLA-A (P = 0.78) (Fig. 7B), HLA-B (P = 0.33) (Fig. 7C), or total HLA-I (P = 0.56) (Fig. 7D). Therefore, transmitted/founder and nontransmitted variants, which varied in their ability to downregulate HLA-C, also varied in the susceptibility to NK cell suppression of virus release.
FIG 7.
Relationship between NK cell suppression and HLA-C downregulation. A Spearman correlation analysis of residual replication in the presence of NK cells against HLA-C (A), HLA-A2 (B), HLA-B7 (C), and total HLA-I (D) downregulation on the x axis. TF (blue) and NT (red) are colored.
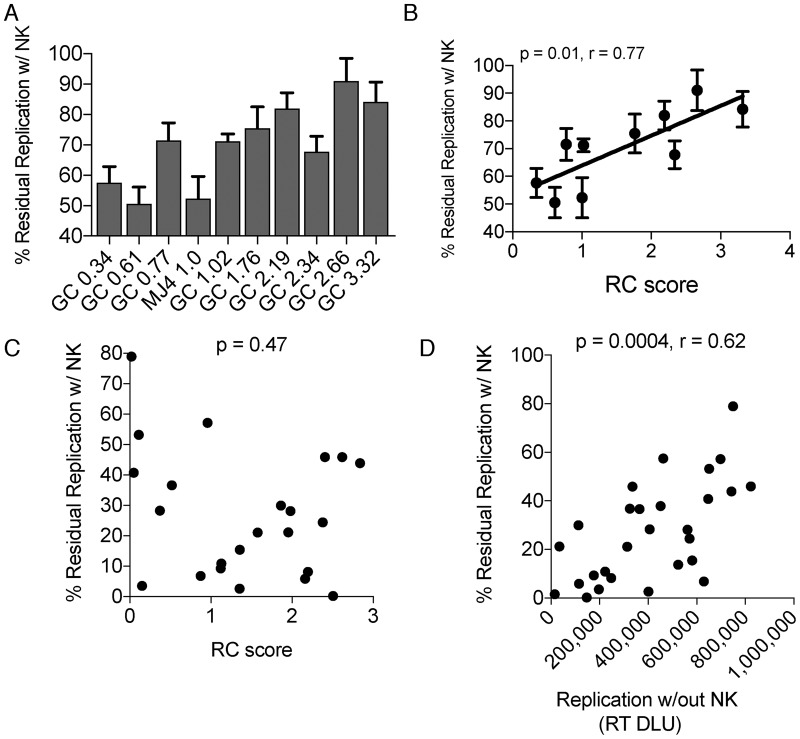
To determine the impact of replicative capacity on NK susceptibility while controlling for variation in HLA-C downregulation, we assessed selected Gag-MJ4 chimeric variants that contain identical Nef and Vpu proteins and exhibit different replicative capacities due to the presence of different Gag genes (49, 50). Gag-MJ4 chimeras were relatively resistant to in vitro NK suppression (Fig. 8A). Susceptibility to NK suppression, as measured by residual replication in the presence of NK cells, correlated with replicative capacity (P = 0.01, r = 0.77) (Fig. 8B). Every 1 unit increase in the replicative capacity score for Gag-MJ4 chimeras (replicative capacity is on a log10 scale) led to an 11% increase in residual replication in the presence of NK cells, indicating an impact of replicative capacity on in vitro NK susceptibility (Fig. 8B). While the replicative capacity of primary variants, as measured previously with a low multiplicity of infection in a spreading assay, did not correlate with NK resistance (P = 0.47) (Fig. 8C), this appeared to be driven primarily by transmission pair 4248, which has a very low RC. In contrast, the levels of virus replication in the absence of NK cells in these high-multiplicity-of-infection experiments did correlate inversely with NK suppression (P = 0.0004, r = 0.62) (Fig. 8D). Thus, the impact of replication extends to primary viruses, although the greater variance observed in Fig. 8D may reflect differences in Vpu that are absent from the Gag-MJ4 chimeric viruses. Normalizing replicative capacity differences of the primary viruses by the measured amount determined from the Gag-MJ4 chimeras (11% increase in residual replication per log increase in RC) did not alter the correlation between HLA-C downregulation and NK suppression of virus release (P = 0.002; r = −0.56) (see Fig. S1).
FIG 8.
Replicative capacity and NK cell suppression. (A) Residual replication in the presence of NK for Gag-MJ4 chimeric viruses (GC) ordered by replicative capacity scores. The x axis contains names of each chimera according to their replicative capacity score, such that variant ID GC 0.34 stands for gag chimera with a replicative capacity score of 0.34. (B and C) Spearman correlation analysis of replicative capacity (RC) versus NK suppression in vitro of Gag-MJ4 chimeras (B) and primary viruses (C). (D) Spearman correlation of residual replication in the presence of NK cells versus replication without NK cells. % of ctrl, percentage of replication in the absence of NK cells.
DISCUSSION
HLA class I downregulation has not been previously investigated in primary cells with well-characterized full-length primary viruses derived from transmission pairs. Here, we investigated HLA class I downregulation in vitro with a previously described panel of 40 subtype C infectious molecular clones derived from near the time of heterosexual transmission. We also evaluated a panel of previously described Gag-MJ4 chimeras to assess the impact of replicative capacity and added the commonly used NL4-3 subtype B laboratory-adapted strain to compare our data to prior studies. We observed variation in HLA class I downregulation by HIV-1 both within and between individuals. HLA-C downregulation, mediated by Vpu, was in general less efficient than that for HLA-B, which was in turn less efficient than that for HLA-A, although perhaps not less impactful on immune recognition, since NK suppression correlated with the capacity to downregulate HLA-C. Despite the selection bias previously observed for consensus residues in the active site of Nef during transmission (38), transmitted/founder variants did not have a common HLA class I downregulation phenotypic signature; rather, they reflected the quasispecies from which they were derived. This is surprising, since even though transmission and establishment of infection may precede the adaptive immune response, one might have expected changes in HLA-I expression to modify susceptibility to the innate immune response.
Downregulation of HLA-A, HLA-B, and HLA-C was common among the primary variants evaluated in this study, and examining all three simultaneously allowed their relationships to be analyzed. Previous studies have observed a relatively tight correlation between HLA-A and HLA-B downregulation in CEM (41) and Jurkat cells (55) using expression vectors and chimeric viruses. We found very similar correlations (Fig. 3B) using authentic viruses in primary cells, which partially validates the use of HLA-A2 expression as a surrogate for HLA-A and HLA-B downregulation for future studies, as was done in the past with cell lines and expression vectors (39, 41, 45, 56–59). Nonetheless, there were differences between HLA-A and HLA-B, as HLA-A2 was generally downregulated more than HLA-B7 (HLA-A/HLA-B ratio, 1.35) (Fig. 3A). This ratio is remarkably similar to what was found previously with primary Nef sequences cloned into NL4-3 in 721.221 cells and HLA-A24 and HLA-B35 downregulation (ratio, ∼1.3) (7). The data presented here do not take into account differences in downregulation that may exist between different HLA-A or HLA-B alleles but are consistent with the notion that HLA-B is more resistant to downregulation than HLA-A, perhaps underpinning the primary role of HLA-B in the control of HIV viremia (7, 60). HLA class I downregulation previously has been measured with a pan-HLA-I antibody (3, 11, 40, 46, 52, 61–65), although fewer studies used primary cells (6, 56, 66). We found total HLA class I correlated with HLA-A2 expression, which is consistent both with a previous study using Jurkat cells (55) and with the fact that surface expression of HLA-A and HLA-B is significantly higher than that of HLA-C (6). Nonetheless, the pan-HLA class I antibody is useful in that it binds to the other HLA-A and HLA-B alleles besides HLA-A2 and HLA-B7. On the other hand, the results are confounded by the fact that they reflect the sum of HLA-A, HLA-B, and HLA-C downregulation, and the extent of HLA-A and HLA-B downregulation did not correlate with that of HLA-C.
A number of amino acid positions in Nef were significantly associated with HLA class I downregulation in our assays, and some of these were found in previous studies (7, 10, 39, 41). However, Vpu sites associated with HLA-C downregulation were not identified by the same analysis. Using upper and lower quartiles instead of terciles gave only one significant Vpu position (position 13). Signatures associated with the pan-HLA class I data yielded three Vpu sites, two of which were toward the N terminus, similar to what was found in the initial study identifying Vpu-mediated HLA-C downregulation (9). Indeed, most of the diversity in the Vpu sequences under study here is in the extracellular and helical transmembrane domain at the N terminus (see Data Set S1 in the supplemental material). Viruses from within pair 3618 that differed substantially in HLA-C downregulation differed most substantially at Vpu positions 2, 4, 18, and 79, although this was not significant. Mutagenesis studies of Nef and Vpu would be needed to validate sites not found in previous studies and specifically to elucidate the mechanism of the interaction of Vpu with HLA-C.
The capacity of primary viruses to downregulate HLA-C correlated directly with in vitro susceptibility to NK suppression, consistent with the very recent results of Körner et al. with the laboratory-adapted strain JRCSF (67). In vitro experiments have shown that HLA-C, when left on the cell surface, can inhibit NK cell killing of HIV-infected cells by engaging inhibitory receptors (5). However, given both the extensive diversity of the NK cell repertoire, which in a single individual includes as many as 30,000 populations of various activating and inhibitory receptor combinations (68), and the fact that other HIV proteins, like Vpr, influence NK cell activation and infected cell killing (69, 70), such a major contribution of a single, albeit crucial, NK ligand is not entirely predictable.
Why is HLA-C downregulation relatively common among primary HIV-1 variants if it leads to NK recognition? The balance between evading NK and CD8 T cells may drive this phenotype. A reasonable hypothesis is that HLA-C downregulation is selected against in acute infection to avoid NK cells and selected for during the adaptive immune response to avoid CD8 T cells, although this is not supported by the data presented here. Transmitted/founder variants were similar in the capacity to downregulate HLA-C (1.3- to 3.1-fold HLA-C downregulation) to the quasispecies from which they were derived and, perhaps as a consequence, were not uniquely resistant to NK suppression. Apps et al. (9) examined nine transmitted/founder variants from subtypes B, C, and D, three of which were subtype C variants, with a 1.4- to 2.5-fold range of HLA-C downregulation in primary cells, suggesting future studies including more transmitted/founder variants will not detect a distinct ability of transmitted/founder variants to downregulate HLA-C and subsequently evade in vitro NK cell suppression. Romani et al. found a similar range of HLA-C downregulation using subtype A Vpu sequences from four patients at various stages of infection (71). If nonspecific NK cell evasion is not contributing to the HIV-1 transmission bottleneck, perhaps specific KIR HLA combinations of each transmission pair play a role, since HIV-1 mutations that lead to evasion of NK cells by peptide-HLA-C stabilization and subsequent binding of inhibitory NK cell ligands has been demonstrated (21, 72). Such a mechanism would necessitate the transmission of preadapted viruses relevant to the recipient's HLA alleles, which is a phenomenon which has been demonstrated in a number of transmission pairs and is a major determinant of viral load and disease progression in recipients (73, 74). However, the impact on NK cells and risk of infection remains unclear, and future studies would need to use in vitro systems that match the KIR and HLA genetics of each transmission pair from whom the viruses were derived.
HLA class I downregulation coincided with peak Gag expression and CD4 downregulation in vitro, while cells with intermediate Gag expression that were CD4+ exhibited little to no HLA-I downregulation (Fig. 2C). Although intermediate Gag expression may represent cells staining positive due to in vitro artifacts (75, 76), this observation is consistent with the results from a study that utilized anti-Gag antibodies and RNA probes to monitor the timing of events that lead to the disruption of surface HLA class I (52). Interestingly, a recent study of SIVMAC239-infected cells ex vivo found little HLA class I downregulation at any stage of the virus life cycle defined by viral RNA transcripts in single cells, even though levels of other surface proteins were altered at different stages (13). Nonetheless, the effect of timing of HLA downregulation indicates the potential to gauge the stage of viral protein expression by alteration of surface receptors, making virus-producing cells indirectly identifiable without cellular fixation and permeabilization (i.e., for live cell sorting, RNA studies, etc.). A study of the precise duration and timing of viral life cycle events coinciding with the alteration of cell surface molecules with primary viruses could benefit therapeutic cure strategies aiming to identify biomarkers associated with the transition from prebursting infected cells to virus-producing cells.
The capacity to downregulate HLA class I was not generally associated with transmission fitness in this study. With the exception of positions 105 and 106 identified in the Sequence Harmony analysis, consensus residues, which are selected for during transmission (35, 38), were not necessarily associated with a particular level of HLA class I downregulation. Additionally, transmitted/founder viruses were not biased toward enhanced or reduced HLA class I (A, B, C, or total) downregulation. Nevertheless, common downregulation signatures often grouped related variants, including transmitted/founder viruses, consistent with the notion that transmitted/founder variants are more often phenotypically closer to their source quasispecies than other transmitted variants. Viral traits that impact disease progression, such as viral load, can be inherited across transmission (74, 77–80), in part due to viral replicative capacity (50). Thus, the heritability of other traits is to be expected. Genetic selection of viruses observed during transmission points to a bias that likely has an associated though undefined phenotype (32, 35, 38, 81–83). However, the magnitude of the predicted phenotypic bias is unclear and is likely to be modest and specific to the circumstances of each transmission event, given the number of factors that influence infection risk and the genetic bottleneck. These include viral load (84, 85), HLA type (22, 23, 86), circumcision status (84, 87, 88), route and direction of transmission (30, 89), presence of genital infections and inflammation (90–92), and the composition of the microbiome (93). For these reasons, the dimensions of the genetic bias may vary significantly depending on the circumstances (38), and this appears to be reflected in the phenotypic data presented here and observed by others (34, 35, 40, 47, 94, 95).
Overall, our results show that quasispecies within and between individuals in the same subtype display a range of HLA class I downregulation capacities that influence immune recognition and may be shifted during transmission to some extent based on the transmitted/founder variant that establishes the new infection, even though no generalizable HLA signature appears to be selected. These data from primary viruses with the full complement of autologous proteins may be important when therapeutic or prophylactic strategies aimed at manipulating CD8 and NK cell responses are being considered.
MATERIALS AND METHODS
Study subjects and samples.
Infectious molecular clones of viruses from 12 subtype C-infected, therapy-naive individuals in six transmission pairs in this study were derived as previously described (35, 51). These individuals come from a heterosexual discordant couple cohort from the Zambia-Emory HIV Research Project who signed informed consent forms agreeing to participate, as were the acutely infected individuals from whom the Gag-MJ4 chimeras were created (50). The University of Zambia Research Ethics Committee and Emory University Institutional Review Board approved the human subject protocols for the couples in this cohort, who were provided monthly counseling and testing prior to the HIV-negative partner becoming positive. All human sample identifier (ID) numbers were anonymized. Epidemiological linkage was determined by HIV-1 gp41 sequences derived from the couple (96), and newly infected recipients were enrolled in the International AIDS Vaccine Initiative (IAVI) Protocol C early infection cohort. Peripheral blood mononuclear cells from a limited number of healthy HIV-negative donors were obtained by leukapheresis by the Emory Center for AIDS Research Clinical Core in collaboration with the Emory Transplant Center by following Emory IRB protocol 45821, “Phlebotomy of healthy donors.”
Phylogenetic tree and sequence alignment.
BioNJ phylogenetic trees from Nef and Vpu amino acid alignments were made using Geneious bioinformatics software (Biomatters, Aukland, New Zealand), ProtTest software (97), and DIVEIN software (98). Full-length Nef and Vpu amino acid sequences were aligned by a MAFTT alignment with Geneious software and then further hand aligned. ProtTest software then was used to derive the best substitution model. The HIVb substitution model was used with 100 bootstraps and the best of NNI and SPR trees for improvements with optimized equilibrium frequencies, an estimated proportion of invariable sites, an estimated gamma distribution, and four substitution rate categories. The trees shown are rooted on the subtype B NL4-3 sequence as an outgroup, and the subtype C consensuses for Nef and Vpu were derived from the Los Alamos National Database (LANL; https://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html). Comparisons in the text to the distance to the full-length subtype C consensus are based on calculations made with 115 full-length concatenated amino acid sequences from these six transmission pairs determined in a prior publication (35). GenBank accession numbers, though previously generated, were not sequential and thus are included in Table S1 in the supplemental material for clarity.
Generation of infectious molecular clones and Gag-MJ4 chimeras.
Subtype C HIV-1 infectious molecular clones (51, 99) and Gag-MJ4 chimeras were described in previous studies (35, 49, 50). Briefly, 40 infectious molecular clones from 12 individuals in 6 transmission pairs from the Zambian cohort were derived from near-full-length single-genome amplifications (SGA) of autologous HIV RNA sequences of infected individuals 22 to 45 days away from the estimated date of transmission (35). Gag-MJ4 chimeras were constructed from patient-derived gag sequences (and 142 nucleotides into pol) of subtype C acutely infected individuals in the Zambian cohort 33 to 49 days away from the estimated date of transmission and cloned into an MJ4 provirus backbone (49, 50, 99), which is a subtype C R5-tropic laboratory strain (100). NL4-3 is a subtype B laboratory strain that is X4 tropic (101). All replication-competent viruses were created by plasmid transfection of 293T cells (American Type Culture Collection). Titers were generated on the TZM-bl indicator cell line (NIH AIDS Research and Reference Reagent Program) to normalize input virus for primary cell infections at a constant multiplicity of infection.
Infections of primary cells in vitro.
Frozen peripheral blood mononuclear cells (PBMC), obtained through Emory IRB protocol 45821 as described above, were thawed in a 37°C water bath and then washed twice in R10 (RPMI 1640 medium supplemented with 1 U/ml penicillin, 10% defined fetal bovine serum [FBS], 1 μg/ml streptomycin, and 300 μg/ml l-glutamine) containing DNase with 1,500-rpm spins for 7 min each. Total PBMC or isolated CD4+ cells (by following the protocol of the EasySep human CD4+ T cell isolation kit; catalog no. 19052) then were stimulated with 2 to 3 μg/ml of phytohemagglutinin (PHA) and 20 U/ml of interleukin-2 (IL-2; reconstituted in phosphate-buffered saline; I7908-10KU; Sigma) in R10 for 48 to 72 h at 2 to 3 million cells per ml in a flask in an incubator at 37°C with 5% CO2 and 95% humidity. Cells then were infected either in 15-ml conical tubes or in 96-well plates at 7 × 105 to 1 × 106 cells per well at a multiplicity of infection of 0.1 to 0.01 by TZM-bl titer from blue cell counts in 250 μl to 300 μl per well. Virus levels in the supernatants were assessed by a radioactive reverse transcriptase (RT) assay described previously and in more detail elsewhere (35, 99), using P33 that detects digital light units (DLU) from a phosphoscreen; thus, RT DLU is reverse transcriptase digital light units. The area under the curve was calculated from mock-subtracted RT DLU values using Prism with a baseline and minimum peak height of zero.
Flow cytometry for HLA downregulation.
Flow cytometry was performed on a BD Fortessa in the Emory Center for AIDS Research Immunology Core. HLA-A2+/HLA-B7+ double-positive primary cells were identified as explained in the text after screening a number of PBMC donors with anti-HLA-A2-Alexa 700 (clone BB7.2; 343317; BioLegend) and anti-HLA-B7-fluorescein isothiocyanate (FITC) (clone BB7.1; MA1-82180; Thermo Scientific) antibodies. One double-positive (HLA-A2+/HLA-B7+), HIV-negative individual was identified, and leukapheresis provided enough cells to perform multiple replicates and experiments.
Infections were done as described above and stained at 7 days postinfection in vitro, which was previously shown to be near the peak of infection for most variants tested here (35). Multiple infection replicates (2 to 4) were pooled for flow cytometry using the following antibodies in multiple experiments: Aqua Live Dead (amine-reactive dye), anti-CD3-allophycocyanin (APC)-Cy7 (SP34.2), anti-CD4-BV711 (OKT4), anti-CD8-QD605 (RPA-T8), anti-HLA-A2-Alexa 700 (BB7.2), anti-HLA-B7-FITC (BB7.1), anti-HLA-C-APC (DT9; conjugated after buffer exchange; Abcam), anti-HLA-ABC-phycoerythrin (PE)-Cy7 (W6/32), and anti-p24Gag-PE (KC57). A median of 2,070 cells (range, 50 to 12,339; NT variant 4473 TF 17 had one replicate pass at 50 infected cells) were analyzed from 2 to 5 independent experiments, with a median of 3.6% infected cells (range, 0.041 to 17%) of live CD3+ CD8− cells. Standard errors calculated from replicates of each variant and averaged for each HLA marker are the following: HLA-A, 0.42; HLA-B, 0.49; HLA-C, 0.16; and total HLA-I, 0.37. Standard deviations calculated the same way are the following: HLA-A, 0.63; HLA-B, 0.75; HLA-C, 0.25; and total HLA-I, 0.56. HLA downregulation was defined as the expression of the HLA marker on singlet+ lymphocyte+ Aqua− CD3+ CD8− CD4+ Gag− cells divided by that of singlet+ lymphocyte+ Aqua− CD3+ CD8− CD4− Gag+ cells. Thus, the difference is a fold change in MFI of a population of cells. No significant differences were observed in CD4 downregulation efficiency.
Genotypic associations with HLA downregulation.
Alignments of amino acid sequences in FASTA format were exported from Geneious software for Sequence Harmony analysis (http://www.ibi.vu.nl/programs/shmrwww/) in order to identify amino acid residues that correlated strongly with class I downregulation. The Nef sequences of viruses within the lowest and highest terciles of HLA-A and HLA-B downregulation were compared, and Vpu sequences were compared similarly for HLA-C downregulation. Concatenated Nef and Vpu sequences were compared for pan-HLA-I downregulation. Sequence Harmony looks for amino acid residues that segregate one group (tercile) from the other, and scores range from 0 to 1, where 1 represents no segregation between two sets of sequences and 0 represents complete segregation of sequences. Z scores represent standard deviations from the mean Sequence Harmony score calculated from 100 random permutations of all sequences; thus, Z scores with the highest negative standard deviation represent the residues most segregated. A Z score of −3 indicates the Sequence Harmony score is three standard deviations below the mean score of 100 permutations. For example, positions where all of the amino acids in one tercile are K and in the other tercile are R will have a Sequence Harmony score of 0 and a very large negative Z score, with the absolute value of the latter depending on the variability and number of sequences analyzed. Significant values with a cutoff of −3 are included in Table 1. Weblogos then were created based on the Sequence Harmony results (http://weblogo.threeplusone.com/create.cgi).
For the estimation of viral adaptation to HLA (data not shown), TF were compared to the median of the variants from donor quasispecies and were compared by a paired Wilcoxon test. The estimation of viral adaptation to an HLA haplotype was performed as previously described (73). Briefly, the degree of adaptation of each viral sequence to the HLA haplotype of the recipient within each transmission pair was defined as the proportion of positions in Gag, Pol, and Nef that could be targeted by each individual according to their HLA alleles that harbor an adapted residue (either nonconsensus or consensus).
NK suppression of replication in vitro.
NK suppression was measured as previously described in He et al., with minor modifications (54). Briefly, isolated CD4 T cells (EasySep Human CD4+ T cell isolation kit) and NK cells (EasySep human NK T cell isolation kit; catalog no. 17955) from the same HIV-negative peripheral blood mononuclear cells were cultured overnight, and CD4 T cells were stimulated with 2 μg/ml PHA and IL-2 (100 U/ml), while NK cells cultured separately were given IL-15 (2 ng/ml reconstituted in phosphate-buffered saline; 247-ILB-005; R&D Systems). Before mixing NK cells (effectors) and CD4 T cells (targets) at various E:T ratios (1:1 to 1:10), 60,000 CD4 T cells were infected at a multiplicity of infection of 0.1 with a spinoculation, or of 1 without spinoculation, in 15-ml conical tubes and then washed with 14 ml RPMI medium and subsequently cultured in V-bottom 96-well plates in 200 μl overnight. The following day, the cells were spun at 2,000 rpm for 5 min, washed twice, and transferred to a round-bottom plate for the addition of NK cells in triplicate in 300 μl total with 50 U/ml IL-2. Supernatants were sampled at days 3, 7, and 11 or just day 7 depending on the experiment. Values reported are from day 7 postinfection and were found to correlate with area under the curve of days 3 to 11 for those where multiple time points were taken. Values shown in the figures have had outliers removed using Prism 7.0 statistical software with the ROUT method and a Q equal to 1% and are median normalized and averaged over 6 replicates to an E:T ratio of 1:10 for consistency. A radioactive reverse transcriptase assay was performed on supernatants to measure the level of virus, and results are reported as DLU, as described previously (35, 99).
HLA and KIR typing.
A detailed description of the HLA and KIR typing can be found in Tang et al. (102) and Merino et al. (28), with the exception that the HLA typing was done using sequencing-based typing without sequence-specific primers and intermediate steps.
Statistics.
Statistics were done using Prism 7 software and JMP Pro 13.0. Nonparametric Spearman correlations are shown in the text and figures as r values. The Wilcoxon matched-pairs signed-rank test was used to compare transmitted/founder variants and matched donor quasispecies medians. Analysis of variance (ANOVA) was used where shown. Linear regressions were performed to compare to historical data and are shown in the text as r2 values. Sequence Harmony analysis was performed to compare amino acids associated with high- and low-level HLA class I downregulation.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge all volunteers and staff at the Zambia Emory HIV Research Project who made this study possible. We thank Cynthia Derdeyn and Kelsie Brooks for thoughtful comments and suggestions on the manuscript; Jon Allen, Sheng Luo, Ling Yue, Paul Farmer, and Dario Dilernia for technical assistance and sample management; and Chris Ibegbu, Kyle Giesler, and Dennis Liota for contributing antibodies during the assay development phase. HLA class I and KIR genotyping was done by Hailin Lu in the laboratory of Jianming Tang at University of Alabama at Birmingham. We thank Barbara Cervasi and Kiran Gil of the Emory Center for AIDS Research Immunology Core for technical assistance, Tianwei Yu of the Emory CFAR Biostatistics Core for expert statistical advice, and Cameron Tran of the Emory CFAR Clinical Core for assistance with obtaining leukapheresed PBMC samples.
This study was funded by R37AI51231 and R01AI64060 (E.H.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was also supported in part by the Yerkes National Primate Research Center base grant ODP51OD11132 through the Office of Research Infrastructure Programs and by the Immunology, Clinical, and Biostatistics Cores of the Emory Center for AIDS Research, supported by grant P30AI050409 from the National Institute of Allergy and Infectious Diseases. M.J.D. was supported in part by a Chateaubriand Fellowship. M.J.D., D.D., and Z.E. were supported in part by Action Cycling Fellowships. E.H. is a Georgia Eminent Scholar.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01633-17.
REFERENCES
- 1.Kerkau T, Schmitt-Landgraf R, Schimpl A, Wecker E. 1989. Downregulation of HLA class I antigens in HIV-1-infected cells. AIDS Res Hum Retrovir 5:613–620. doi: 10.1089/aid.1989.5.613. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz O, Maréchal V, Le Gall S, Lemonnier F, Heard JM. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med 2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 3.Le Gall S, Erdtmann L, Benichou S, Berlioz-Torrent C, Liu L, Benarous R, Heard JM, Schwartz O. 1998. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8:483–495. doi: 10.1016/S1074-7613(00)80553-1. [DOI] [PubMed] [Google Scholar]
- 4.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 5.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671. doi: 10.1016/S1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 6.Apps R, Meng Z, Del Prete GQ, Lifson JD, Zhou M, Carrington M. 2015. Relative expression levels of the HLA class-I proteins in normal and HIV-infected cells. J Immunol 194:3594–3600. doi: 10.4049/jimmunol.1403234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahiti M, Toyoda M, Jia X, Kuang XT, Mwimanzi F, Mwimanzi P, Walker BD, Xiong Y, Brumme ZL, Brockman MA, Ueno T. 2016. Relative resistance of HLA-B to downregulation by naturally occurring HIV-1 Nef sequences. mBio 7:e01516-. doi: 10.1128/mBio.01516-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Specht A, DeGottardi MQ, Schindler M, Hahn B, Evans DT, Kirchhoff F. 2008. Selective downmodulation of HLA-A and -B by Nef alleles from different groups of primate lentiviruses. Virology 373:229–237. doi: 10.1016/j.virol.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Apps R, Del Prete GQ, Chatterjee P, Lara A, Brumme ZL, Brockman MA, Neil S, Pickering S, Schneider DK, Piechocka-Trocha A, Walker BD, Thomas R, Shaw GM, Hahn BH, Keele BF, Lifson JD, Carrington M. 2016. HIV-1 Vpu mediates HLA-C downregulation. Cell Host Microbe 19:686–695. doi: 10.1016/j.chom.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis MJ, Lee P, Ng HL, Yang OO. 2012. Immune selection in vitro reveals human immunodeficiency virus type 1 Nef sequence motifs important for its immune evasion function in vivo. J Virol 86:7126–7135. doi: 10.1128/JVI.00878-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swigut T, Alexander L, Morgan J, Lifson J, Mansfield KG, Lang S, Johnson RP, Skowronski J, Desrosiers R. 2004. Impact of Nef-mediated downregulation of major histocompatibility complex class I on immune response to simian immunodeficiency virus. J Virol 78:13335–13344. doi: 10.1128/JVI.78.23.13335-13344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich TC, Piaskowski SM, Leon EJ, Furlott JR, Maness NJ, Weisgrau KL, Mac Nair CE, Weiler AM, Loffredo JT, Reynolds MR, Williams KY, Klimentidis YC, Wilson NA, Allison DB, Rakasz EG. 2010. High viremia is associated with high levels of in vivo major histocompatibility complex class I downregulation in rhesus macaques infected with simian immunodeficiency virus SIVmac239. J Virol 84:5443–5447. doi: 10.1128/JVI.02452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton DL, McGinnis K, Finak G, Chattopadhyay P, Gottardo R, Roederer M. 2017. Combined single-cell quantitation of host and SIV genes and proteins ex vivo reveals host-pathogen interactions in individual cells. PLoS Pathog 13:e1006445. doi: 10.1371/journal.ppat.1006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. 2007. Cutting edge: allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol 178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 15.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. 2007. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med 204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31:429–434. [DOI] [PubMed] [Google Scholar]
- 17.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrington M, Martin MP, van Bergen J. 2008. KIR-HLA intercourse in HIV disease. Trends Microbiol 16:620–627. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blish CA. 2016. Natural killer cell diversity in viral infection: why and how much? Pathog Immun 1:165–192. doi: 10.20411/pai.v1i1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, Del Prete GQ, Goulder P, Brumme ZL, Brumme CJ, John M, Mallal S, Nelson G, Bosch R, Heckerman D, Stein JL, Soderberg KA, Moody MA, Denny TN, Zeng X, Fang J, Moffett A, Lifson JD, Goedert JJ, Buchbinder S, Kirk GD, Fellay J, McLaren P, Deeks SG, Pereyra F, Walker B, Michael NL, Weintrob A, Wolinsky S, Liao W, Carrington M. 2013. Influence of HLA-C expression level on HIV control. Science 340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, Carrington M, Allen TM, Altfeld M. 2011. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorak MT, Tang J, Penman-Aguilar A, Westfall AO, Zulu I, Lobashevsky ES, Kancheya NG, Schaen MM, Allen SA, Kaslow RA. 2004. Transmission of HIV-1 and HLA-B allele-sharing within serodiscordant heterosexual Zambian couples. Lancet 363:2137–2139. doi: 10.1016/S0140-6736(04)16505-7. [DOI] [PubMed] [Google Scholar]
- 23.Mackelprang RD, John-Stewart G, Carrington M, Richardson B, Rowland-Jones S, Gao X, Mbori-Ngacha D, Mabuka J, Lohman-Payne B, Farquhar C. 2008. Maternal HLA homozygosity and mother-child HLA concordance increase the risk of vertical transmission of HIV-1. J Infect Dis 197:1156–1161. doi: 10.1086/529528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koehler RN, Alter G, Tovanabutra S, Saathoff E, Arroyo MA, Walsh AM, Sanders-Buell EE, Maboko L, Hoelscher M, Robb ML, Michael NL, McCutchan FE, Kim JH, Kijak GH. 2013. Natural killer cell-mediated innate sieve effect on HIV-1: the impact of KIR/HLA polymorphism on HIV-1 subtype-specific acquisition in east Africa. J Infect Dis 208:1250–1254. doi: 10.1093/infdis/jit349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boulet S, Kleyman M, Kim JY, Kamya P, Sharafi S, Simic N, Bruneau J, Routy J-P, Tsoukas CM, Bernard NF. 2008. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 22:1487–1491. doi: 10.1097/QAD.0b013e3282ffde7e. [DOI] [PubMed] [Google Scholar]
- 26.Jennes W, Verheyden S, Mertens JW, Camara M, Seydi M, Dieye TN, Mboup S, Demanet C, Kestens L. 2013. Inhibitory KIR/HLA incompatibility between sexual partners confers protection against HIV-1 transmission. Blood 121:1157–1164. doi: 10.1182/blood-2012-09-455352. [DOI] [PubMed] [Google Scholar]
- 27.Jennes W, Verheyden S, Demanet C, Adjé-Touré CA, Vuylsteke B, Nkengasong JN, Kestens L. 2006. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol 177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 28.Merino A, Malhotra R, Morton M, Mulenga J, Allen S, Hunter E, Tang J, Kaslow RA. 2011. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J Infect Dis 203:487–495. doi: 10.1093/infdis/jiq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandler NG, Bosinger SE, Estes JD, Zhu RTR, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, Del Prete GQ, Hill BJ, Timmer JK, Reiss E, Yarden G, Darko S, Contijoch E, Todd J-P, Silvestri G, Nason M, Norgren RB, Keele BF, Rao S, Langer JA, Lifson JD, Schreiber G, Douek DC. 2014. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boily M-C, Baggaley RF, Wang L, Mâsse B, White RG, Hayes RJ, Alary M. 2009. Heterosexual risk of HIV-1 infection per sexual act: systematic review and meta-analysis of observational studies. Lancet Infect Dis 9:118–129. doi: 10.1016/S1473-3099(09)70021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw GM, Hunter E. 2012. HIV transmission. Cold Spring Harb Perspect Med 2:a006965. doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joseph SB, Swanstrom R, Kashuba ADM, Cohen MS. 2015. Bottlenecks in HIV-1 transmission: insights from the study of founder viruses. Nat Rev Microbiol 13:414–425. doi: 10.1038/nrmicro3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer SS, Bibollet-Ruche F, Sherrill-Mix S, Learn GH, Plenderleith L, Smith AG, Barbian HJ, Russell RM, Gondim MVP, Bahari CY, Shaw CM, Li Y, Decker T, Haynes BF, Shaw GM, Sharp PM, Borrow P, Hahn BH. 2017. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc Natl Acad Sci U S A 114:E590–E599. doi: 10.1073/pnas.1620144114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberle CS, Joos B, Rusert P, Campbell NK, Beauparlant D, Kuster H, Weber J, Schenkel CD, Scherrer AU, Magnus C, Kouyos R, Rieder P, Niederöst B, Braun DL, Pavlovic J, Böni J, Yerly S, Klimkait T, Aubert V, Trkola A, Metzner KJ, Günthard HF, Swiss HIV Cohort Study (SHCS). 2016. Tracing HIV-1 transmission: envelope traits of HIV-1 transmitter and recipient pairs. Retrovirology 13:62. doi: 10.1186/s12977-016-0299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deymier MJ, Ende Z, Fenton-May AE, Dilernia DA, Kilembe W, Allen SA, Borrow P, Hunter E. 2015. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-α resistance. PLoS Pathog 11:e1005154. doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster TL, Wilson H, Iyer SS, Coss K, Doores K, Smith S, Kellam P, Finzi A, Borrow P, Hahn BH, Neil SJD. 2016. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe 20:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin T-H, Peng J, Seese AM, Shapiro R, Frater J, Ndung'u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJR, Allen TM, Allen S, Hunter E. 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345:1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mann JK, Chopera D, Omarjee S, Kuang XT, Le AQ, Anmole G, Danroth R, Mwimanzi P, Reddy T, Carlson J, Radebe M, Goulder PJR, Walker BD, Abdool Karim S, Novitsky V, Williamson C, Brockman MA, Brumme ZL, Ndung'u T. 2014. Nef-mediated down-regulation of CD4 and HLA class I in HIV-1 subtype C infection: association with disease progression and influence of immune pressure. Virology 468-470:214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mlcochova P, Apolonia L, Kluge SF, Sridharan A, Kirchhoff F, Malim MH, Sauter D, Gupta RK. 2015. Immune evasion activities of accessory proteins Vpu, Nef and Vif are conserved in acute and chronic HIV-1 infection. Virology 482:72–78. doi: 10.1016/j.virol.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann JK, Byakwaga H, Kuang XT, Le AQ, Brumme CJ, Mwimanzi P, Omarjee S, Martin E, Lee GQ, Baraki B, Danroth R, McCloskey R, Muzoora C, Bangsberg DR, Hunt PW, Goulder PJR, Walker BD, Harrigan PR, Martin JN, Ndung'u T, Brockman MA, Brumme ZL. 2013. Ability of HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes. Retrovirology 10:100. doi: 10.1186/1742-4690-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuang XT, Li X, Anmole G, Mwimanzi P, Shahid A, Le AQ, Chong L, Qian H, Miura T, Markle T, Baraki B, Connick E, Daar ES, Jessen H, Kelleher AD, Little S, Markowitz M, Pereyra F, Rosenberg ES, Walker BD, Ueno T, Brumme ZL, Brockman MA. 2014. Impaired Nef function is associated with early control of HIV-1 viremia. J Virol 88:10200–10213. doi: 10.1128/JVI.01334-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mwimanzi P, Markle TJ, Ueno T, Brockman MA. 2012. Human leukocyte antigen (HLA) class I down-regulation by human immunodeficiency virus type 1 negative factor (HIV-1 Nef): what might we learn from natural sequence variants? Viruses 4:1711–1730. doi: 10.3390/v4091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mwimanzi P, Markle TJ, Martin E, Ogata Y, Kuang XT, Tokunaga M, Mahiti M, Pereyra F, Miura T, Walker BD, Brumme ZL, Brockman MA, Ueno T. 2013. Attenuation of multiple Nef functions in HIV-1 elite controllers. Retrovirology 10:1. doi: 10.1186/1742-4690-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shahid A, Olvera A, Anmole G, Kuang XT, Cotton LA, Plana M, Brander C, Brockman MA, Brumme ZL. 2015. Consequences of HLA-B*13-associated escape mutations on HIV-1 replication and Nef function. J Virol 89:11557–11571. doi: 10.1128/JVI.01955-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahiti M, Brumme ZL, Jessen H, Brockman MA, Ueno T. 2015. Dynamic range of Nef-mediated evasion of HLA class II-restricted immune responses in early HIV-1 infection. Biochem Biophys Res Commun 463:248–254. doi: 10.1016/j.bbrc.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 47.Noviello CM, Pond SLK, Lewis MJ, Richman DD, Pillai SK, Yang OO, Little SJ, Smith DM, Guatelli JC. 2007. Maintenance of Nef-mediated modulation of major histocompatibility complex class I and CD4 after sexual transmission of human immunodeficiency virus type 1. J Virol 81:4776–4786. doi: 10.1128/JVI.01793-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis MJ, Balamurugan A, Ohno A, Kilpatrick S, Ng HL, Yang OO. 2008. Functional adaptation of Nef to the immune milieu of HIV-1 infection in vivo. J Immunol 180:4075–4081. doi: 10.4049/jimmunol.180.6.4075. [DOI] [PubMed] [Google Scholar]
- 49.Claiborne DT, Prince JL, Scully E, Macharia G, Micci L, Lawson B, Kopycinski J, Deymier MJ, Vanderford TH, Nganou-Makamdop K, Ende Z, Brooks K, Tang J, Yu T, Lakhi S, Kilembe W, Silvestri G, Douek D, Goepfert PA, Price MA, Allen SA, Paiardini M, Altfeld M, Gilmour J, Hunter E. 2015. Replicative fitness of transmitted HIV-1 drives acute immune activation, proviral load in memory CD4+ T cells, and disease progression. Proc Natl Acad Sci U S A 112:E1480–E1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prince JL, Claiborne DT, Carlson JM, Schaefer M, Yu T, Lahki S, Prentice HA, Yue L, Vishwanathan SA, Kilembe W, Goepfert P, Price MA, Gilmour J, Mulenga J, Farmer P, Derdeyn CA, Tang J, Heckerman D, Kaslow RA, Allen SA, Hunter E. 2012. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog 8:e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deymier MJ, Claiborne DT, Ende Z, Ratner HK, Kilembe W, Allen S, Hunter E. 2014. Particle infectivity of HIV-1 full-length genome infectious molecular clones in a subtype C heterosexual transmission pair following high fidelity amplification and unbiased cloning. Virology 468-470C:454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martrus G, Niehrs A, Cornelis R, Rechtien A, García-Beltran W, Lütgehetmann M, Hoffmann C, Altfeld M. 2016. Kinetics of HIV-1 latency reversal quantified on the single-cell level using a novel flow-based technique. J Virol 90:9018–9028. doi: 10.1128/JVI.01448-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brandt BW, Feenstra KA, Heringa J. 2010. Multi-harmony: detecting functional specificity from sequence alignment. Nucleic Acids Res 38:W35–W40. doi: 10.1093/nar/gkq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He X, Simoneau CR, Granoff ME, Lunemann S, Dugast A-S, Shao Y, Altfeld M, Körner C. 2016. Assessment of the antiviral capacity of primary natural killer cells by optimized in vitro quantification of HIV-1 replication. J Immunol Methods 434:53–60. doi: 10.1016/j.jim.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Specht A, Telenti A, Martinez R, Fellay J, Bailes E, Evans DT, Carrington M, Hahn BH, Goldstein DB, Kirchhoff F. 2010. Counteraction of HLA-C-mediated immune control of HIV-1 by Nef. J Virol 84:7300–7311. doi: 10.1128/JVI.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hung C-H, Thomas L, Ruby CE, Atkins KM, Morris NP, Knight ZA, Scholz I, Barklis E, Weinberg AD, Shokat KM, Thomas G. 2007. HIV-1 Nef assembles a Src family kinase-ZAP-70/Syk-PI3K cascade to downregulate cell-surface MHC-I. Cell Host Microbe 1:121–133. doi: 10.1016/j.chom.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Schaefer MR, Wonderlich ER, Roeth JF, Leonard JA, Collins KL. 2008. HIV-1 Nef targets MHC-I and CD4 for degradation via a final common beta-COP-dependent pathway in T cells. PLoS Pathog 4:e1000131. doi: 10.1371/journal.ppat.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasper MR, Collins KL. 2003. Nef-mediated disruption of HLA-A2 transport to the cell surface in T cells. J Virol 77:3041–3049. doi: 10.1128/JVI.77.5.3041-3049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ali A, Realegeno S, Yang OO, Lewis MJ. 2009. Simultaneous assessment of CD4 and MHC-I downregulation by Nef primary isolates in the context of infection. J Virol Methods 161:297–304. doi: 10.1016/j.jviromet.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajapaksa US, Li D, Peng Y-C, McMichael AJ, Dong T, Xu X-N. 2012. HLA-B may be more protective against HIV-1 than HLA-A because it resists negative regulatory factor (Nef) mediated down-regulation. Proc Natl Acad Sci U S A 109:13353–13358. doi: 10.1073/pnas.1204199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamada T, Kaji N, Odawara T, Chiba J, Iwamoto A, Kitamura Y. 2003. Proline 78 is crucial for human immunodeficiency virus type 1 Nef to down-regulate class I human leukocyte antigen. J Virol 77:1589–1594. doi: 10.1128/JVI.77.2.1589-1594.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mangasarian A, Piguet V, Wang JK, Chen YL, Trono D. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol 73:1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X, Schrager JA, Lange GD, Marsh JW. 2001. HIV Nef-mediated cellular phenotypes are differentially expressed as a function of intracellular Nef concentrations. J Biol Chem 276:32763–32770. doi: 10.1074/jbc.M101025200. [DOI] [PubMed] [Google Scholar]
- 64.Akari H, Arold S, Fukumori T, Okazaki T, Strebel K, Adachi A. 2000. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J Virol 74:2907–2912. doi: 10.1128/JVI.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galaski J, Ahmad F, Tibroni N, Pujol FM, Müller B, Schmidt RE, Fackler OT. 2016. Cell surface downregulation of NK cell ligands by patient-derived HIV-1 Vpu and Nef alleles. J Acquir Immune Defic Syndr 72:1–10. doi: 10.1097/QAI.0000000000000917. [DOI] [PubMed] [Google Scholar]
- 66.Marodon G, Landau NR, Posnett DN. 1999. Altered expression of CD4, CD54, CD62L, and CCR5 in primary lymphocytes productively infected with the human immunodeficiency virus. AIDS Res Hum Retrovir 15:161–171. doi: 10.1089/088922299311583. [DOI] [PubMed] [Google Scholar]
- 67.Körner C, Simoneau CR, Schommers P, Granoff M, Ziegler M, Hölzemer A, Lunemann S, Chukwukelu J, Corleis B, Naranbhai V, Kwon DS, Scully EP, Jost S, Kirchhoff F, Carrington M, Altfeld M. 2017. HIV-1-mediated downmodulation of HLA-C impacts target cell recognition and antiviral activity of NK cells. Cell Host Microbe 22:111–119.e4. doi: 10.1016/j.chom.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, Davis MM, Norman PJ, Guethlein LA, Desai M, Parham P, Blish CA. 2013. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 5:208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richard J, Sindhu S, Pham TNQ, Belzile J-P, Cohen EA. 2010. HIV-1 Vpr up-regulates expression of ligands for the activating NKG2D receptor and promotes NK cell-mediated killing. Blood 115:1354–1363. doi: 10.1182/blood-2009-08-237370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ward J, Davis Z, DeHart J, Zimmerman E, Bosque A, Brunetta E, Mavilio D, Planelles V, Barker E. 2009. HIV-1 Vpr triggers natural killer cell-mediated lysis of infected cells through activation of the ATR-mediated DNA damage response. PLoS Pathog 5:e1000613. doi: 10.1371/journal.ppat.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romani B, Kavyanifard A, Allahbakhshi E. 2017. Functional conservation and coherence of HIV-1 subtype A Vpu alleles. Sci Rep 7:87. doi: 10.1038/s41598-017-00222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fadda L, Körner C, Kumar S, van Teijlingen NH, Piechocka-Trocha A, Carrington M, Altfeld M. 2012. HLA-Cw*0102-restricted HIV-1 p24 epitope variants can modulate the binding of the inhibitory KIR2DL2 receptor and primary NK cell function. PLoS Pathog 8:e1002805. doi: 10.1371/journal.ppat.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monaco DC, Dilernia DA, Fiore-Gartland A, Yu T, Prince JL, Dennis KK, Qin K, Schaefer M, Claiborne DT, Kilembe W, Tang J, Price MA, Farmer P, Gilmour J, Bansal A, Allen S, Goepfert P, Hunter E. 2016. Balance between transmitted HLA preadapted and nonassociated polymorphisms is a major determinant of HIV-1 disease progression. J Exp Med 213:2049–2063. doi: 10.1084/jem.20151984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carlson JM, Du VY, Pfeifer N, Bansal A, Tan VYF, Power K, Brumme CJ, Kreimer A, DeZiel CE, Fusi N, Schaefer M, Brockman MA, Gilmour J, Price MA, Kilembe W, Haubrich R, John M, Mallal S, Shapiro R, Frater J, Harrigan PR, Ndung'u T, Allen S, Heckerman D, Sidney J, Allen TM, Goulder PJR, Brumme ZL, Hunter E, Goepfert PA. 2016. Impact of pre-adapted HIV transmission. Nat Med 22:606–613. doi: 10.1038/nm.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heynen CA, Holzer TJ. 1992. Evaluation of a flow cytometric model for monitoring HIV antigen expression in vitro. J Immunol Methods 152:25–33. doi: 10.1016/0022-1759(92)90085-8. [DOI] [PubMed] [Google Scholar]
- 76.Terahara K, Yamamoto T, Mitsuki Y-Y, Shibusawa K, Ishige M, Mizukoshi F, Kobayashi K, Tsunetsugu-Yokota Y. 2011. Fluorescent reporter signals, EGFP, and DsRed, encoded in HIV-1 facilitate the detection of productively infected cells and cell-associated viral replication levels. Front Microbiol 2:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fraser C, Lythgoe K, Leventhal GE, Shirreff G, Hollingsworth TD, Alizon S, Bonhoeffer S. 2014. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343:1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yue L, Prentice HA, Farmer P, Song W, He D, Lakhi S, Goepfert P, Gilmour J, Allen S, Tang J, Kaslow RA, Hunter E. 2013. Cumulative impact of host and viral factors on HIV-1 viral-load control during early infection. J Virol 87:708–715. doi: 10.1128/JVI.02118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lingappa JR, Thomas KK, Hughes JP, Baeten JM, Wald A, Farquhar C, de Bruyn G, Fife KH, Campbell MS, Kapiga S, Mullins JI, Celum C, Partners in Prevention HSV/HIV Transmission Study Team. 2013. Partner characteristics predicting HIV-1 set point in sexually acquired HIV-1 among African seroconverters. AIDS Res Hum Retrovir 29:164–171. doi: 10.1089/aid.2012.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hollingsworth TD, Laeyendecker O, Shirreff G, Donnelly CA, Serwadda D, Wawer MJ, Kiwanuka N, Nalugoda F, Collinson-Streng A, Ssempijja V, Hanage WP, Quinn TC, Gray RH, Fraser C. 2010. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog 6:e1000876. doi: 10.1371/journal.ppat.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 82.Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol 79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 84.Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, Meehan MO, Lutalo T, Gray RH. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N Engl J Med 342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 85.Fideli US, Allen SA, Musonda R, Trask S, Hahn BH, Weiss H, Mulenga J, Kasolo F, Vermund SH, Aldrovandi GM. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retrovir 17:901–910. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang X, Ling H, Mao W, Ding X, Zhou Q, Han M, Wang F, Cheng L, Xiong H. 2009. Association of HLA-A, B, DRB1 alleles and haplotypes with HIV-1 infection in Chongqing, China. BMC Infect Dis 9:201. doi: 10.1186/1471-2334-9-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Auvert B, Taljaard D, Lagarde E, Sobngwi-Tambekou J, Sitta R, Puren A. 2005. Randomized, controlled intervention trial of male circumcision for reduction of HIV infection risk: the ANRS 1265 trial. PLoS Med 2:e298. doi: 10.1371/journal.pmed.0020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gray RH, Kigozi G, Serwadda D, Makumbi F, Watya S, Nalugoda F, Kiwanuka N, Moulton LH, Chaudhary MA, Chen MZ, Sewankambo NK, Wabwire-Mangen F, Bacon MC, Williams CFM, Opendi P, Reynolds SJ, Laeyendecker O, Quinn TC, Wawer MJ. 2007. Male circumcision for HIV prevention in men in Rakai, Uganda: a randomised trial. Lancet 369:657–666. doi: 10.1016/S0140-6736(07)60313-4. [DOI] [PubMed] [Google Scholar]
- 89.Baggaley RF, White RG, Boily M-C. 2010. HIV transmission risk through anal intercourse: systematic review, meta-analysis and implications for HIV prevention. Int J Epidemiol 39:1048–1063. doi: 10.1093/ije/dyq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, Hahn BH, Allen SA, Derdeyn CA, Hunter E. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog 5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Masson L, Passmore J-AS, Liebenberg LJ, Werner L, Baxter C, Arnold KB, Williamson C, Little F, Mansoor LE, Naranbhai V, Lauffenburger DA, Ronacher K, Walzl G, Garrett NJ, Williams BL, Couto-Rodriguez M, Hornig M, Lipkin WI, Grobler A, Abdool Karim Q, Abdool Karim SS. 2015. Genital inflammation and the risk of HIV acquisition in women. Clin Infect Dis 61:260–269. doi: 10.1093/cid/civ298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cameron DW, Simonsen JN, D'Costa LJ, Ronald AR, Maitha GM, Gakinya MN, Cheang M, Ndinya-Achola JO, Piot P, Brunham RC. 1989. Female to male transmission of human immunodeficiency virus type 1: risk factors for seroconversion in men. Lancet ii:403–407. [DOI] [PubMed] [Google Scholar]
- 93.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, Dong M, Chen Y, Ismail N, Ndung'u T, Ghebremichael MS, Wesemann DR, Mitchell C, Dong KL, Huttenhower C, Walker BD, Virgin HW, Kwon DS. 2017. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity 46:29–37. doi: 10.1016/j.immuni.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Etemad B, Gonzalez OA, White L, Laeyendecker O, Kirk GD, Mehta S, Sagar M. 2014. Characterization of HIV-1 envelopes in acutely and chronically infected injection drug users. Retrovirology 11:106. doi: 10.1186/s12977-014-0106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog 8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, Kasolo F, Musonda R, Hunter E, Gao F, Allen S, Hahn BH. 2002. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol 76:397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 98.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI. 2010. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques 48:405–408. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Claiborne DT, Prince JL, Hunter E. 2014. A restriction enzyme based cloning method to assess the in vitro replication capacity of HIV-1 subtype C Gag-MJ4 chimeric viruses. J Vis Exp 2014:51506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ndung'u T, Renjifo B, Essex M. 2001. Construction and analysis of an infectious human Immunodeficiency virus type 1 subtype C molecular clone. J Virol 75:4964–4972. doi: 10.1128/JVI.75.11.4964-4972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tang J, Malhotra R, Song W, Brill I, Hu L, Farmer PK, Mulenga J, Allen S, Hunter E, Kaslow RA. 2010. Human leukocyte antigens and HIV type 1 viral load in early and chronic infection: predominance of evolving relationships. PLoS One 5:e9629. doi: 10.1371/journal.pone.0009629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.