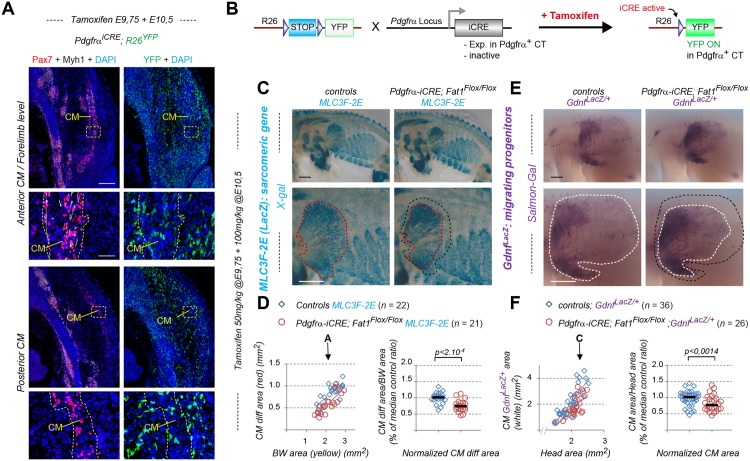
Fig 9. Inducible Fat1 deletion in the pdgfrα connective tissue lineage alters progression of CM migration and differentiation.
(A) Cross sections of a Pdgfrα-iCre; R26YFP/+ embryo collected at E12.5, after in utero administration of tamoxifen at E9.75 (50 mg/kg) + E10.5 (100 mg/kg). Alternate sections at anterior and posterior CM levels, respectively, were immunostained with antibodies against: left panels: Pax7 (red) and myh1 (white), plus DAPI (blue); right panels: GFP (green), plus DAPI (blue), to reveal the outcome of Pdgfrα-iCre-mediated R26-YFP recombination (right panels). The yellow dotted boxes indicate the areas magnified in the bottom panels. (B) Principle of the genetic paradigm used to follow the Pdgfrα-iCre lineage, using the R26Lox-STOP-Lox-YFP reporter line combined with Pdgfrα-iCre. iCRE (CRE/ERT2) is expressed in the domain of Pdgfrα expression but remains catalytically inactive. iCRE activity is permitted by in utero treatment with tamoxifen. Catalytic activity is triggered in the cells expressing iCRE at the time of tamoxifen treatment, thus allowing the stop cassette to be deleted and YFP to be permanently expressed. (C, E) Whole-mount β-galactosidase staining was performed using X-gal as substrate on embryos carrying the MLC3F-2E transgene (C) or using Salmon-Gal as substrate on embryos carrying the GdnfLacZ/+ allele (E) in the context of tamoxifen-induced Fat1 deletion in the Pdgfrα lineage driven by Pdgfrα-iCre at E12.5. Top images show a side view of the whole flank of an embryo. Lower images are higher magnification of the area in which the CM spreads. Red and white dotted lines correspond to the areas covered in control embryos by MLC3F-2E+ CM fibers (C) and by GdnfLacZ+ progenitors (E), respectively. In comparison, the corresponding areas observed in mutants are indicated as black dotted lines in both cases. (D) Quantification of the expansion rate of differentiated CM fibers. Left graph: For each embryo side, the area covered by differentiated CM fibers was plotted relative to the area occupied by body wall muscles. Right plot: for each embryo, the CM area/body wall area was normalized to the median ratio of control embryos. Blue dots: Fat1Flox/Flox; MLC3F-2E (n = 22, control embryos from tamoxifen-treated litters); red dots: Pdgfrα-iCre; Fat1Flox/Flox- ; MLC3F-2E (n = 21). Underlying data are provided in S1 Data. (F) Quantification of the expansion rate of the area occupied by GdnfLacZ+ progenitors. Left plot: for each embryo side, the area covered by GdnfLacZ+ progenitors was plotted relative to the trunk length. Right plot: for each embryo, the GdnfLacZ+ CM area/trunk length was normalized to the median ratio of control embryos. Blue dots: Fat1Flox/Flox; GdnfLacZ/+ (n = 36; control embryos from tamoxifen-treated litters); red dots: Pdgfrα-iCre; Fat1Flox/Flox-; GdnfLacZ/+ (n = 26). Underlying data are provided in S1 Data. Scale bars: (A) low magnification: 200 μm; high magnification: 50 μm; (C, E) 500 μm. CM, cutaneous maximus; CRE/ERT2, CRE fused with the estrogen receptor Tamoxifen-binding domain; iCre, short form of CRE/ERT2; Pdgfrα, platelet-derived growth factor receptor alpha.