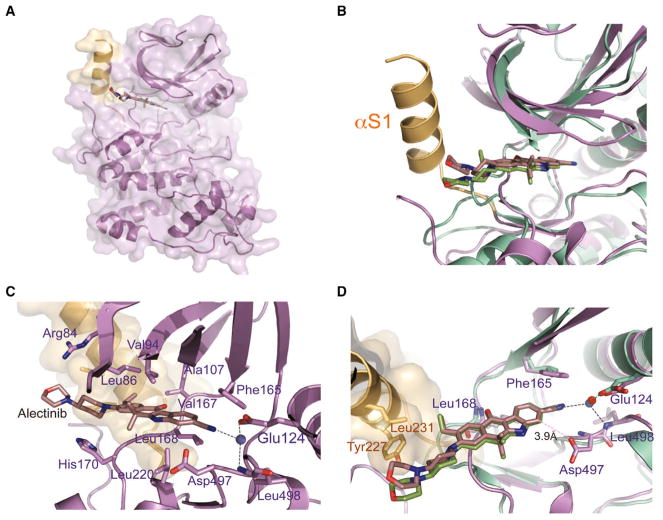
Figure 2. Crystal Structure of the SRPK1-Alectinib Complex.
(A) Overall structure of SRPK1 in complex with Alectinib. Helix αS1 of the non-conserved spacer region forms a continuous molecular surface adjacent to the ATP-binding pocket and is colored light orange.
(B) Comparison of the SRPK1-Alectinib and ALK-Alectinib complex structures. The two structures were superimposed using the carbons of their small lobes. SRPK1-Alectinib complex and ALK-Alectinib complex are colored purple and green, respectively. The unique helix αS1 of SRPK1 is colored light orange.
(C) Alectinib forms an extensive network of interactions with both the small and large lobes of SRPK1. Residues that interact with Alectinib are indicated.
(D) Alectinib hydrogen bonds with the backbone amide of Leu168 of SRPK1 (purple). A crucial solvent water molecule (blue) mediates a network of hydrogen bonds between Alectinib, the side chain of Glu124 and the backbone amide of Leu498 of SRPK1. All hydrogen bonds are denoted by black dotted lines. van der Waals interaction between Asp497 and Alectinib is denoted by pink dotted line. Extra contacts between the 4-morpholinopiperidine moiety of Alectinib and the side chains of Tyr227 and Leu231 of SRPK1 are observed. The structure of ALK-Alectinib complex is shown in green for comparison. The crucial water molecule in the ALK-Alectinib structure is colored red.
See also Table S2.