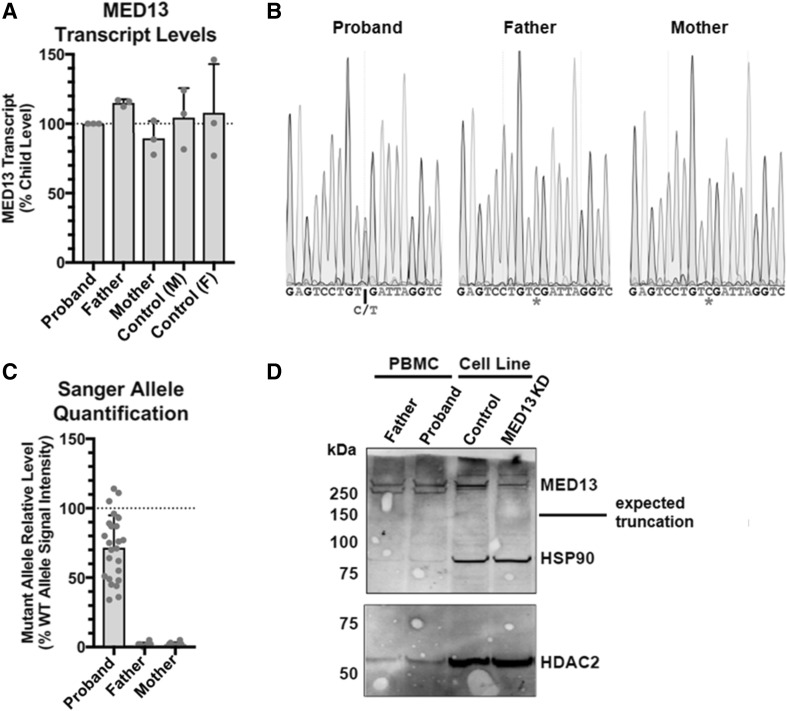
Fig. 4.
Analysis of transcript and protein levels in patient with nonsense mutation. a Level of MED13 transcript was measured by qPCR and normalized to GAPDH and proband (patient J). No differences were detectable between groups (One-way ANOVA p = 0.5913). An additional loading control (AGPAT) produced very similar results (data not shown). b Representative Sanger traces from cDNA amplicons demonstrating the presence of the variant in the proband, and absence in the father and mother. c Quantification of the chromatograms of all Sanger sequences reveals less signal from the base on the mutant allele (p < 0.0001 by paired t-test compared to the wildtype base signal by trace). The father and mother do not have any signal at the mutant base above the level of noise. d Western blot for MED13 (and HSP90 and HDAC2 as loading controls) from nuclear extracts of patient peripheral blood mononuclear cells or a neural precursor cell line (present to demonstrate antibody specificity with a knockdown (KD) control). If the nonsense mutation resulted in a stable protein, a product at approximately 150 kDa would be expected, which is not present. No protein was recoverable from the blood sample from the mother