Figure 1.
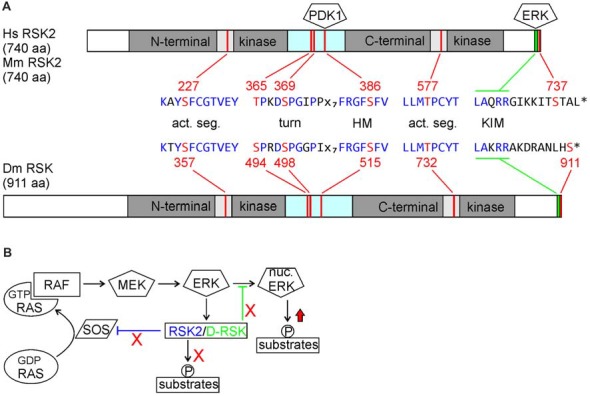
(A) Comparison of human and mouse ribosomal S6 kinase 2 (RSK2) with Drosophila D-RSK. Conservation of all relevant phosphorylation sites (red) embedded in common consensus sequences (blue) indicates a common mode of activation. Activated ERK binds to the C-terminal kinase interaction motif (KIM) and induces catalytic activity of the C-terminal kinase domain (CTKD) by phosphorylation of a threonine residue (RSK2: T577; D-RSK: T732) in the kinase activation segment. The CTKD in turn phosphorylates a serine residue (RSK2: S386; D-RSK: S515) in the hydrophobic motif (HM) located in the linker region, which promotes binding and activation of 3-phosphoinositde-dependent kinase 1 (PDK1). Furthermore, ERK phosphorylates two residues (RSK2: T365, S369; D-RSK: S494, S498) in the turn motif next to the N-terminal kinase domain (NTKD). In combination with PDK1-mediated phosphorylation of serine S227 (D-RSK: S357) this stabilizes the active conformation of the NTKD, as shown for other AGC-type kinases (Leroux et al., 2018). Involvement of Drosophila PDK1 in D-RSK activation was deduced from genetic interaction studies (Rintelen et al., 2001). Release of ERK is promoted by NTKD-mediated autophosphorylation of serine 737. Whether the corresponding C-terminal serine residue 911 in D-RSK has a similar function is not known. (B) Integration of RSK2 and D-RSK in MAP-kinase signaling. Loss-of-function mutations not only abolish phosphorylation of RSK substrate proteins but also prevent feedback inhibition (red crosses) resulting in enhanced ERK-mediated phosphorylation of substrate proteins (red arrow). Different mechanisms for negative regulation of ERK by RSK2 (inhibition of RAS activation) or D-RSK (inhibition of ERK nuclear translocation) have been described.
