Figure 1.
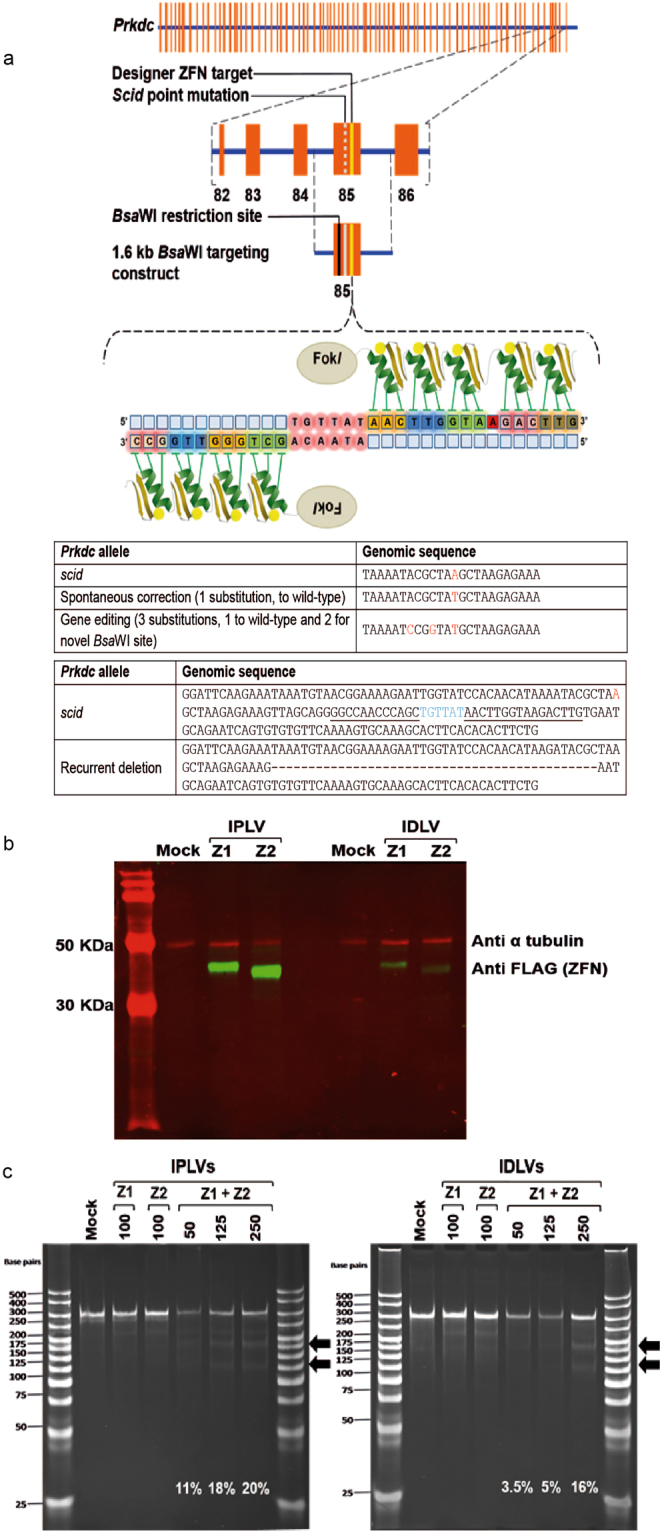
Prkdc gene editing strategy and outcomes, ZFN design and gene expression, and cutting activity in scid fibroblasts. (a) Schematic of Prkdc, including location of scid and ZFN target sites, structure of ZFN and sequences of various Prkdc alleles around scid site. (b) Production of ZFN monomers from SFFV-driven IPLV and IDLV (qPCR MOI 100). Proteins were extracted 3 d after transduction. The blotting revealed FLAG-tagged ZFN monomers (42 and 38.5 kDa for ZFN1 & ZFN2, respectively), with α-tubulin used as a loading control (49 kDa). (c) Cel-I analysis of Prkdc cutting by ZFN. Scid fibroblasts were transduced with IPLV- and IDLV-ZFN at the indicated MOI and genomic DNA was extracted 3 d later. The Cel-I-digested PCR products were separated on a polyacrylamide gel and diagnostic bands (black arrows) were used to calculate the % of indels induced by the various ZFN treatments and indicated on the gel image.
