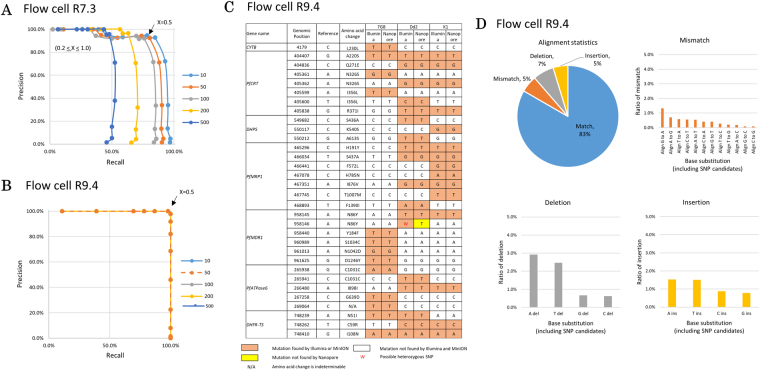
Figure 2.
SNP calling of laboratory strains. The precision and recall of multiple thresholds used for heterozygous SNP calling on data obtained with flow cell R7.3. Precision is defined as the ratio of true positive (i.e., SNPs found by both MinION and Illumina) to the total of true positive and false positive (i.e., SNPs found only by MinION). Recall is defined as the ratio of true positive to the total of true positive and false negative (i.e., SNPs found only by Illumina). The lines show the sequence depth used for analysis (threshold R). The dots on the line show the threshold X. Threshold X ≥ 0.5 with R ≥ 50 sequence depth is used for subsequent analysis because of the reasonable precision and recall values, which are 0.92 and 0.8, respectively (A). On the other hand, flow cell R9.4 gives the same precision and recall for every threshold R with X ≥ 0.5, which is 1 and 0.97, respectively. In this figure, all the lines produced by different threshold R stack up (B). SNPs called with the parameters X ≥ 0.5 and R ≥ 50 on data obtained with flow cell R9.4 show 100% identity to Illumina validation (orange boxes). Exception is made for one position in PfMDR1 (red color font). This position is seemingly heterozygous as called by Illumina. However, MinION could only determine this position as a homozygous SNP (C). The error patterns show that G to A mismatches, A deletions, and A/T insertions are the most frequent polymorphisms in the laboratory strain malaria parasites sequenced with flow cell R9.4 (D).