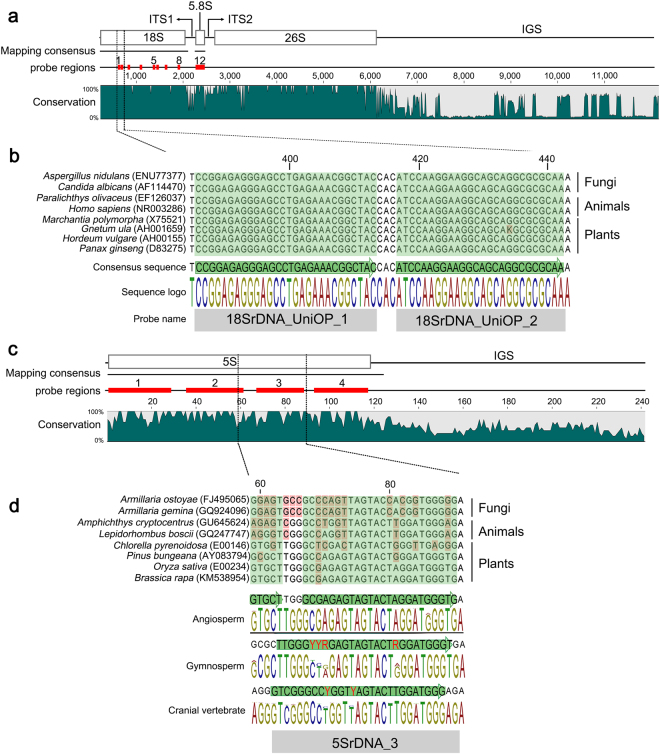
Figure 1.
Design of the 45S and 5S rDNA PLOPs. (a) Diagram of the 45S rDNA unit of P. ginseng (KM036296), as commonly observed in most plant species. The consensus region was generated from mapping sequences downloaded from public databases (Supplementary Fig. S1). The 12 PLOP regions for 45S rDNA are indicated by red bars. The sequence conservation for each region of the 45S rDNA is represented by homologous sequence depth across 32 complete rDNA-IGS from different species. (b) Two examples of PLOPs in the 18S region of 45S rDNA. All sequences are conserved across different fungal, animal, and plant taxa. The bottom part of the figure shows the consensus sequence, sequence logo, and probe names. (c) Diagram of the 5S rDNA unit of P. ginseng (KM036312), as commonly observed in most plant species. The four 5S rDNA PLOP regions are indicated by red bars, and the sequence conservation is represented as the depth of homologous sequences across eight species. (d) An example of PLOPs based on 5S rDNA. The 5S rDNA coding regions are highly polymorphic across distantly related taxa. To address this issue, different PLOPs were designed for each lineage of angiosperm, gymnosperm, and cranial vertebrate species (see Table 1). The consensus sequence, sequence logo, and probe name are shown. Degenerate nucleotides (red nucleotides in the consensus sequences) were incorporated into the probes (thick green arrows) to include polymorphic regions, and each type was pooled to form the “universal” 5S rDNA FISH probe cocktail.