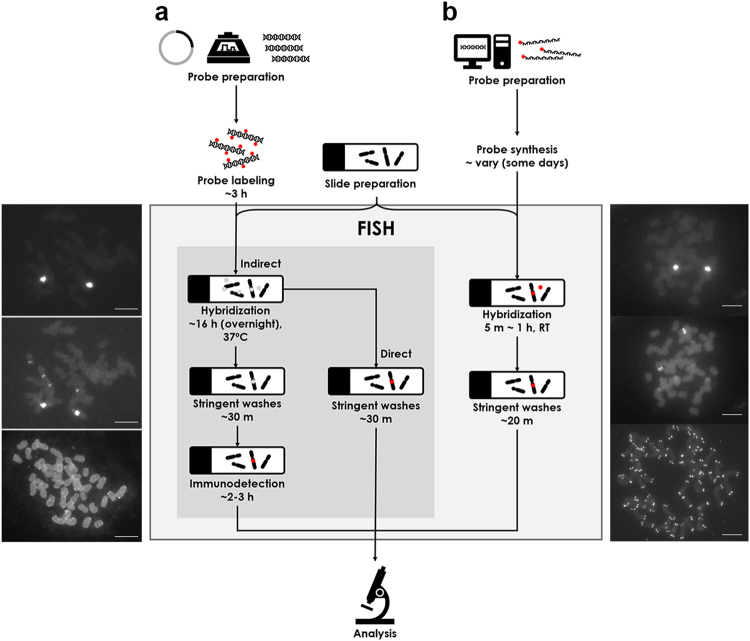
Figure 3.
Schematic diagram comparing the conventional FISH and PLOP-FISH workflows. (a) Probe preparation prior to FISH may involve cloning or PCR amplification of a target sequence and subsequent nick-translation labeling with either haptens (indirect) or fluorochromes (direct), producing ~200–500 bp probes. FISH using these relatively long double-stranded DNA probes involves overnight hybridization at 37 °C, and if the probe is labeled with a hapten, immunodetection should be carried out. Conventional FISH using indirectly labeled probes by nick-translation typically involves additional steps such as RNase and protease treatment prior to the hybridization reaction (not shown in diagram) and an immunodetection step. Probes directly labeled with fluorochromes may or may not be treated with RNase and protease. Both methods need overnight hybridization, making them more time consuming and labor intensive than PLOP-FISH. (b) PLOP-FISH begins with the design of probes from bioinformatically analyzed sequences to optimize probe length (~30 bp) and melting temperature (~45–50 °C at 2 × SSC and 50% formamide) for rapid hybridization at room temperature. Ordering of fluorochrome-prelabeled oligonucleotide sequences eliminates the labeling step but may take some days to be delivered. Probe preparation for both conventional and PLOP-FISH may vary depending on the nature of probe source. The general processes and number of steps required for PLOP-FISH are reduced compared with conventional FISH, thus reducing the chances for error, while simplifying and expediting downstream analyses. Inset images in the left and right panels show conventional and PLOP-FISH signals of 45S rDNA, 5S rDNA, and telomere (top to bottom), respectively. Bars = 10 µm.