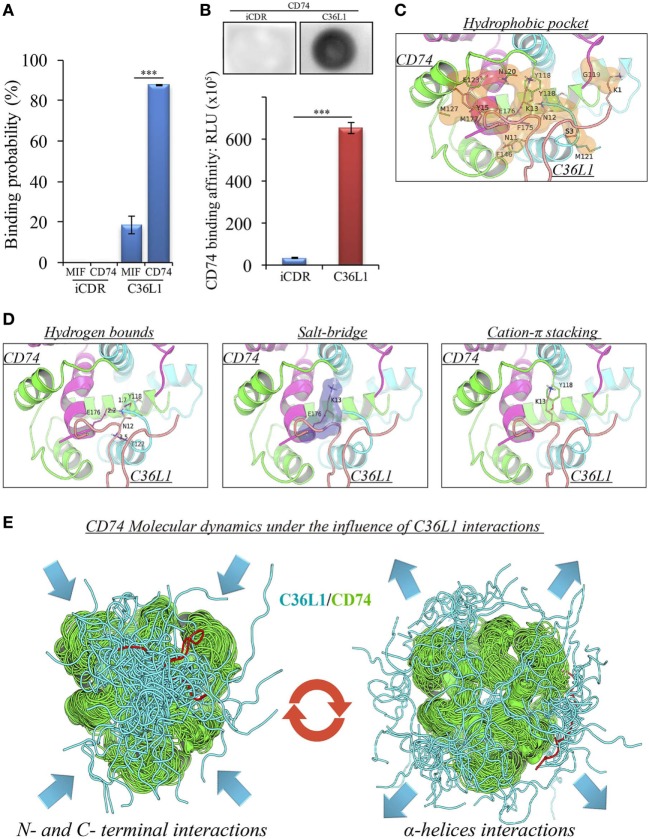
Figure 4.
Binding prediction and molecular docking of C36L1 dynamic interactions to macrophage migration inhibitory factor (MIF) and its receptor CD74. (A) Binding probability of C36L1 peptide and irrelevant peptide (iCDR) to MIF and its receptor CD74 calculated using PepSite algorithm. Best ranked binding scores (n = 5) were included in the analysis for each group (***p < 0.001). (B) Dot-blot binding assay for C36L1 and iCDR peptides to mouse recombinant CD74. Bar graph represents mean of RLU in dot area quantified using ImageJ software from triplicates (n = 3), ***p < 0.001. (C) Hydrophobic pocket (orange) formed by CD74 and C36L1 partners characterized by carbon–carbon interactions above a 4 Å distance cutoff. (D) Electrostatic interactions between CD74 and C36L1 peptide: hydrogen bonds formed between partners. Donor–acceptor distances are described; salt bridge formed involving K13; cation–π stacking between tyrosine residues of chain A of CD74 and C36L1. CD74 chains A, B, and C are colored in green, cyan, and magenta, respectively. C36L1 is colored in yellow. (E) Overlap of highest and lowest free energy results for C36L1 (cyan) in complex with CD74 (green). Left: Overlap of the lowest free energy 50 poses showing major concentration of C36L1 peptide at the CD74 N- and C-terminal interface. Lowest peptide free energy pose highlighted in red. Right: Overlap of the lowest free energy 50 poses, where C36L1 visits other regions of CD74, including the external region of the α-helices. Lowest peptide free energy highlighted in red.