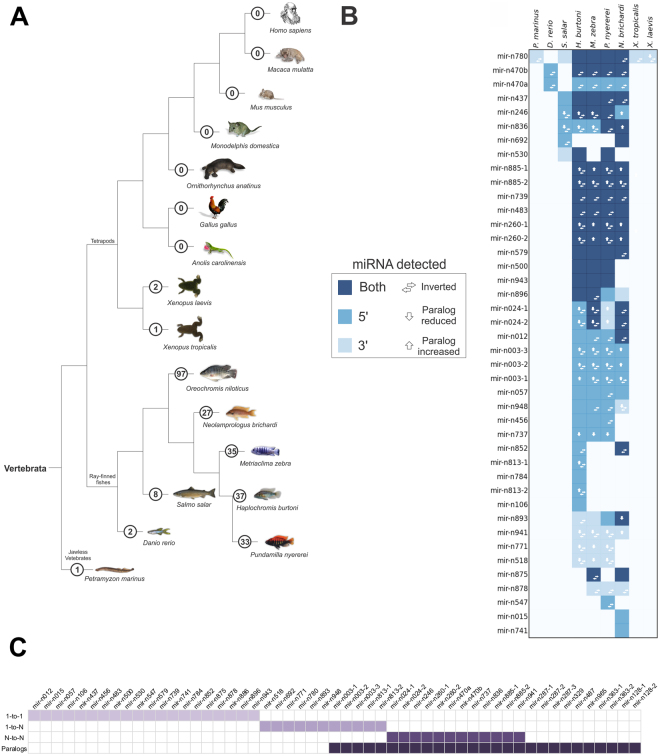
Figure 8.
Detection of novel Nile tilapia miRNAs in diverse vertebrate species by miRDeep2 approach and ortholog/paralog annotation. (A) Vertebrate evolutionary tree with the number of novel miRNAs (inside circles) detected in each species through genome-wide matching searches by miRDeep2 software. Phylogenetic tree is a handmade tree derived by merging published trees by Brawand et al.39 and Amemiya et al.94. Homo sapiens, Ornithorhynchus anatinus, Petromyzon marinus, Danio rerio, Salmo salar, Gallus gallus and Monodelphis domestica images were obtained from https://commons.wikimedia.org/wiki/Main_Page under the Public Domain; Macaca mulata and Mus musculus images from https://pixabay.com under the Public Domain; Pundamilia nyererei, Haplochromis burtoni and Neolamprologus brichardi adapted by permission from Macmillan Publishers Ltd: Nature (Brawand et al.39), copyright 2014, https://www.nature.com/; Xenopus tropicalis and Xenopus laevis images from http://journals.plos.org/plosbiology/ under Creative Commons Attribution 4.0 International (CC BY 4.0 - authors “Patrick Narbonne, David E. Simpson and John B. Gurdon”, title “Figure S1. Characterization of the hybrids formed by the cross-fertilization of X. laevis eggs with X. tropicalis sperm.”, date “15 November, 2011”, url “https://doi.org/10.1371/journal.pbio.1001197.s001”, no endorsement, only esthetical adaptations: image vectorized, background removed, shadow included and image rotated); Metriaclima zebra or Maylandia zebra and Anolis carolinensis images from http://www.publicdomainpictures.net under CC0 1.0 Universal: CC0 1.0 Public Domain Dedication; Oreochromis niloticus image from https://www.usgs.gov/ under the Public domain. (B) Comparative features of 42 novel miRNAs among species, including sense orientation, paralogs abundance, and arms detection. Other 55 novel miRNAs were tilapia-specific, and therefore, excluded from this comparative analysis. (C) Orthologs and paralogs relationship cardinalities considering common anscestral and the “young” gene copies from novel Nile tilapia miRNA genes. Orthologs were infered from reconciled gene and species trees obtained with RAxML and TreeBest using predicted data and homologs miRNA data from miRBase (see methodology).