Figure 3.
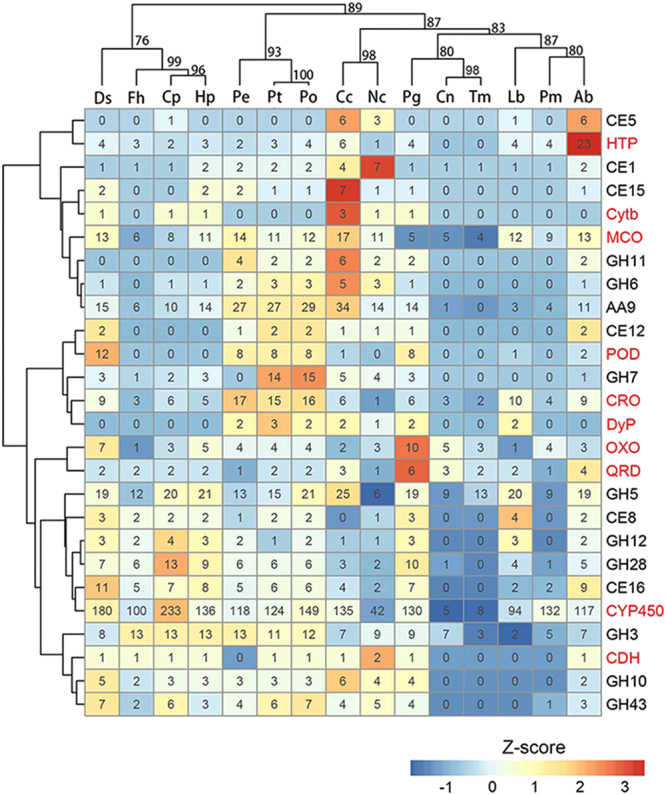
Clustered gene contents of CAZyme and oxidoreductase families in fifteen representative fungi genomes. Hierarchical clustering of wood-decay related genes were completed using the R package pvclust, in which the unbiased (AU) p-values (%) are computed by 1000 bootstrap re-samplings. Respective gene numbers of each gene family in corresponding genome is shown in each cell. Over-represented and under-represented gene families are depicted in red and blue backgrounds, respectively. Within the gene family on the right column, abbreviated names of CAZyme and lignolytic oxidoreductase families are denoted in black and red, respectively. Abbreviations of gene family names: GH, glycoside hydrolases; CE, carbohydrate esterases; POD, class II peroxidases; MCO, multicopper oxidases; CRO, copper-radical oxidases; CDH, cellobiose dehydrogenase; Cytb562, cytochrome b562; OXO, oxalate oxidase/decarboxylases; QRD, quinone reductases; DyP, dye-decolorizing peroxidases; HTP, heme-thiolate peroxidases; P450, cytochromes P450. Abbreviations of species names: Po, Pleurotus ostreatus; Pt, Pleutotus tuoliensis; Pe, Pleurotus eryngii var. eryngii; Fh, Fistulina hepatica; Ab, Agaricus bisporus; Lb, Laccaria bicolor; Cc, Coprinopsis cinerea; Cp, Coniophora puteana; Hp, Hydnomerulius pinastri; Pm, Pisolithus microcarpus; Ds, Dichomitus squalens; Pg, Phlebiopsis gigantean; Cn, Cryptococcus neoformans; Tm, Tremella mesenterica; Nc, Neurospora crassa.
