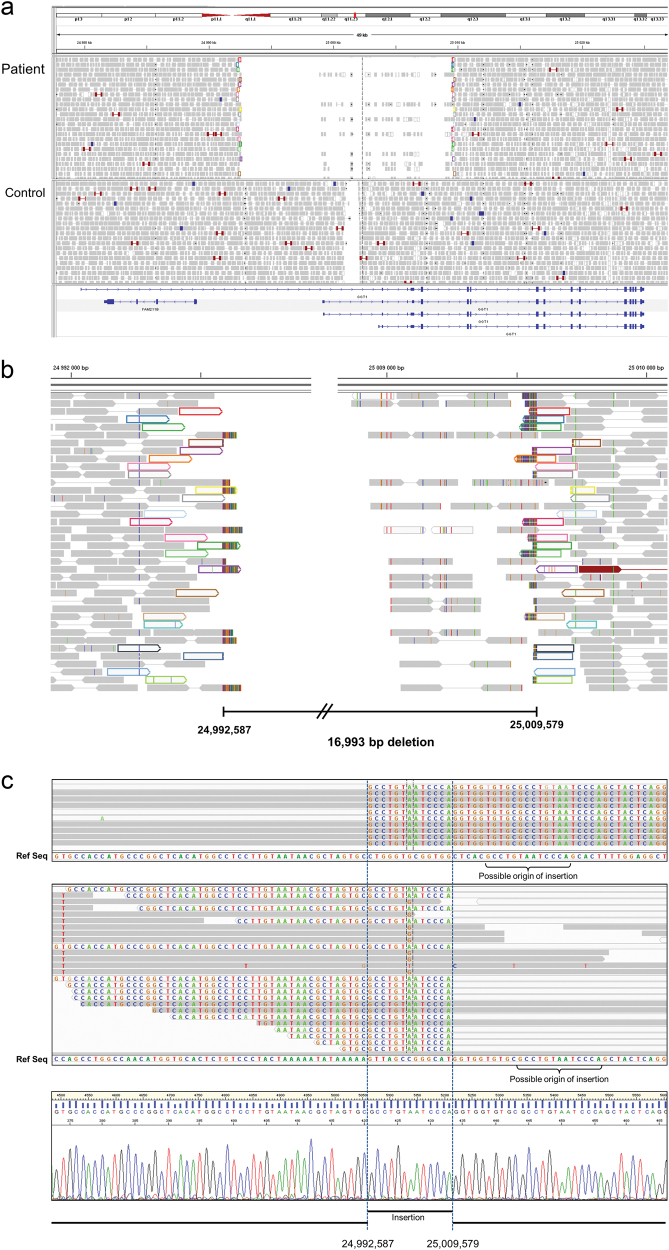
Fig. 3.
Visualization of WGS data covering GGT1. a Plot of paired reads from patient 1 indicates the presence of a homozygous deletion in GGT1. The deletion includes the first coding exon of all isoforms of GGT1 as well as several non-coding exons. All control samples tested presented a low coverage region that corresponds to a low complexity DNA sequence. b Mapping of soft-clipped ends defines the deletion between positions Chr22:24,992,587 and Chr22:25,009,579. Paired reads flanking the deletion are depicted in matching colours. c. Alignment of soft-clipped ends from the centromeric breakpoint (upper panel) and telomeric breakpoint (middle panel) reveals the additional presence of a 13 bp duplication. Sanger sequencing of the deletion junction (lower panel) confirms the WGS findings