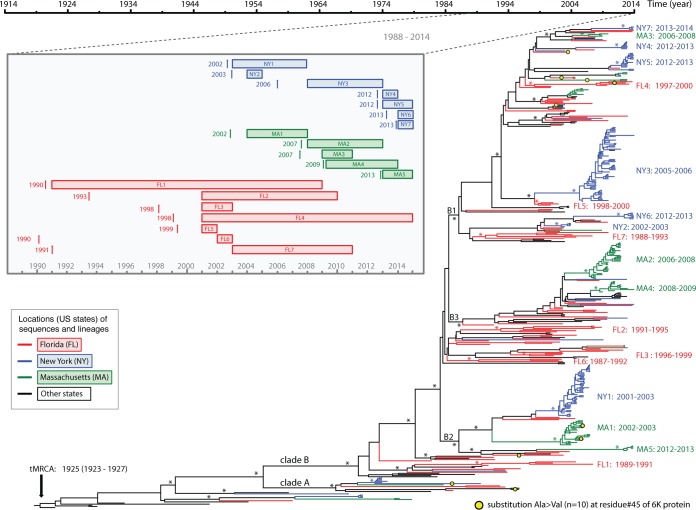
FIG 3.
Time-scaled phylogeny of EEEV. A maximum clade credibility (MCC) tree of complete genome sequences of EEEV is shown. Major nodes with posterior probabilities over 0.9 are marked by asterisks. Sequences from Florida, New York, Massachusetts, and other places are colored and described in the legend key. Clades A and B and subclades B1, B2, and B3, as well as the 19 monophyletic groups further defined for Florida, New York, and Massachusetts viruses, are shown. The times of the most recent common ancestors (tMRCAs) of all EEEVs and the monophyletic groups are provided after the colons. Mutations at residue 45 (Ala→Vla) in the 6K protein are mapped on the branches of the tree using yellow circles. The inset diagram on the left shows the time spans of these monophyletic groups, including the range of the sampling time (rectangular boxes) and the mean of the tMRCAs (vertical arrow bars).