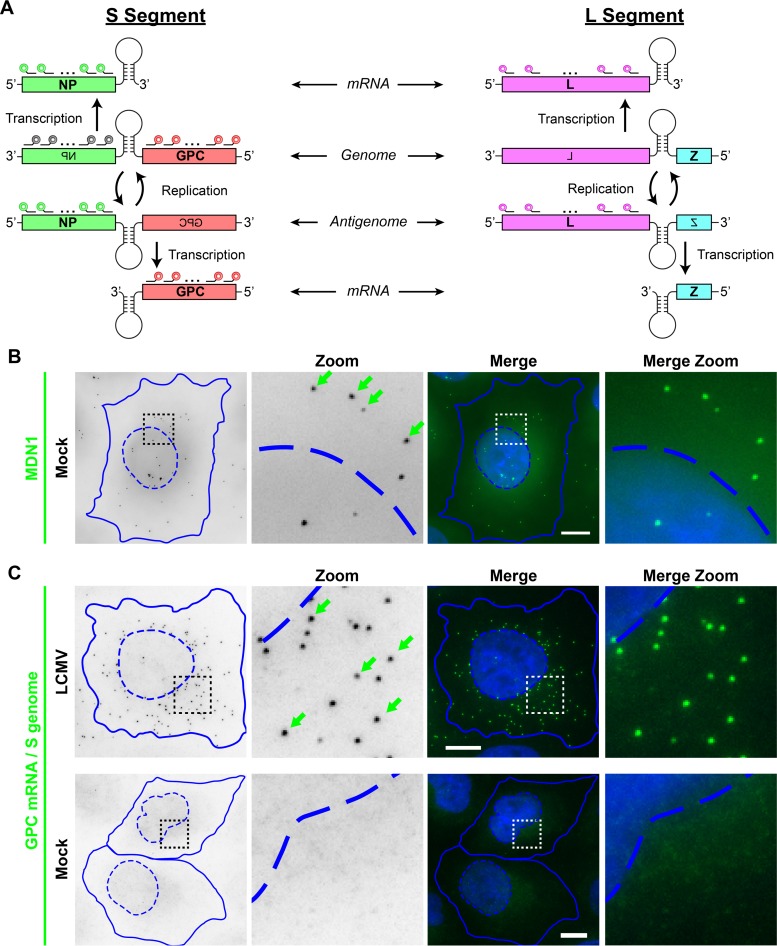
FIG 1.
LCMV RNA species can be specifically visualized by using multiple, singly labeled oligonucleotide smFISH probes. (A) Overview of the scheme used by arenaviruses to transcribe and replicate their single-stranded, ambisense, bisegmented genome. smFISH probes that recognize the S segment genomic RNA are shown in gray, probes that recognize the S segment genome and GPC mRNA are shown in red, probes that recognize the S segment antigenome and NP mRNA are shown in green, and probes that recognize the L segment antigenome and L mRNA are shown in pink. smFISH probe sets consist of pools of 48 individual 20-mer oligonucleotides, each labeled with a single fluorophore at their 3′ termini. (B) Uninfected cells were stained with a control smFISH probe set specific for MDN1 cellular mRNA labeled with Cy3. (C) Cells either were infected with LCMV at an MOI of 0.01 or, as a control, remained uninfected (mock). Cells were fixed at 24 hpi and stained with a Cy5-labeled smFISH probe set specific for S segment genomic RNA and GPC mRNA. Boxed regions of the cell are magnified and shown in columns labeled “Zoom.” Green arrows indicate example smFISH-stained spots most likely representing single labeled RNAs. Nuclear (hatched lines) and cytoplasmic (solid lines) boundaries are shown in blue. The same intensity levels for a particular probe set were applied to all images of mock- and LCMV-infected cells to permit comparisons. Bars, 10 μm.