FIG 1.
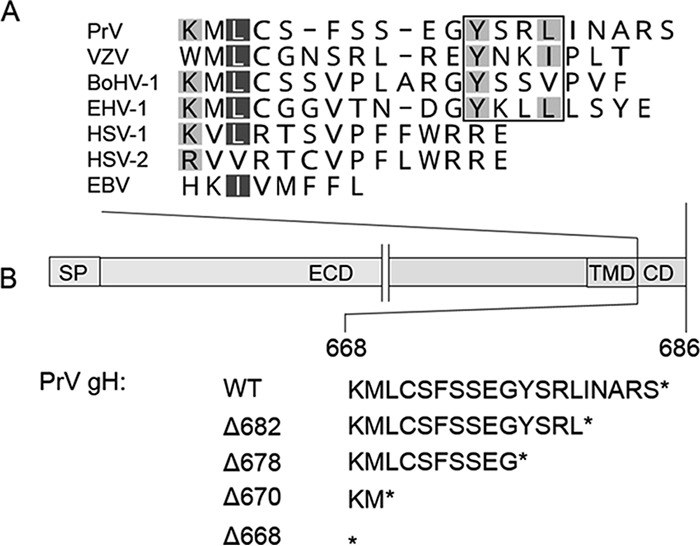
Sequence alignment of gH cytoplasmic domains of different herpesviruses and schematic representation of the generated gH mutants. (A) The alignment was generated using ClustalW (Geneious 10.3.2). Residues are colored according to their conservation (white letters on dark gray background, 80 to 100%; black letters on light gray, 60 to 80%). The YXXϕ endocytosis motif is boxed. Sequences of herpesvirus gH used for the alignment correspond to gH of pseudorabies virus (PrV; AAA47466), varicella-zoster virus (VZV; AH010537.2), bovine alphaherpesvirus 1 (BoHV-1; CAA41677), equid alphaherpesvirus 1 (EHV-1; AY464052), human alphaherpesvirus 1 (HSV-1; AJE60179), human alphaherpesvirus 2 (HSV-2; CAB06746), and Epstein-Barr virus (EBV; V01555.2). (B) The PrV gH open reading frame and the amino acid sequence of the cytoplasmic domains of the generated truncation mutants are shown. SP, signal peptide; ECD, ectodomain; TMD, transmembrane domain; CD, cytoplasmic domain.
