Figure 1. BFG‐GI pipeline summary.
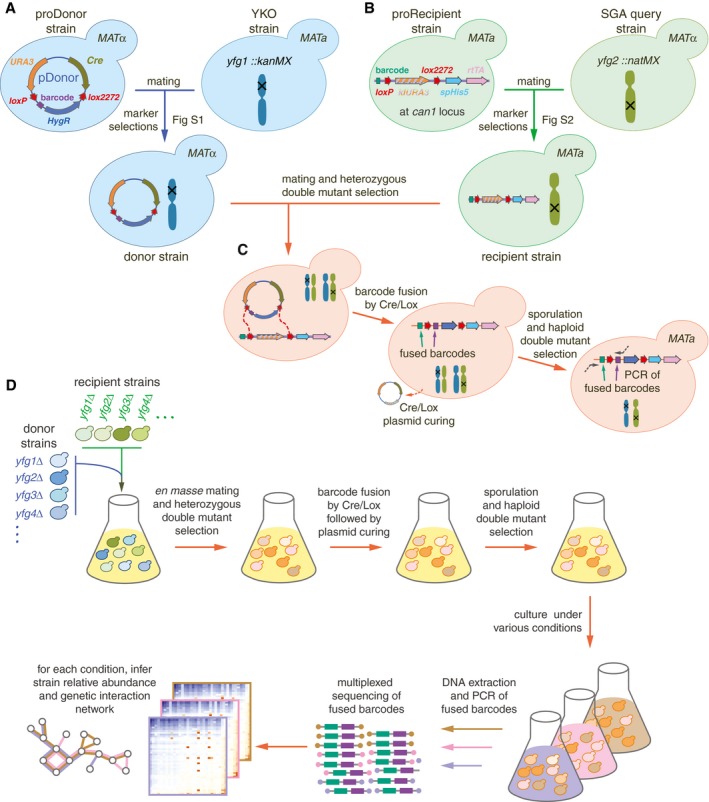
- Construction of donors with unique barcodes representing each gene deletion in parental strains from the YKO collection.
- Construction of recipients also with unique barcodes representing genes of interest in parental strains from the SGA query collection. Pairs of recombination sites (loxP and lox2272) were located at the barcode loci of donor and recipient strains to enable in vivo intracellular fusion of barcode pairs at the recipient barcode locus.
- Donors and recipients were mated with each other to generate heterozygous diploid double mutants, and barcodes were fused in vivo by the Cre/Lox system. The relic plasmid remaining in donors after Cre/Lox recombination was counter‐selected after barcode fusion. Sporulation was induced to select for the MAT a progeny and haploid double mutants.
- BFG‐GI was conducted en masse to generate “many‐by‐many” pools for a set of 26 DNA repair and 14 neutral genes. The resulting pool of haploid double mutants was stored as aliquots of glycerol stock. Thawed aliquots were used to inoculate media containing different chemical agents (“drugs”). Genomic DNA was extracted and fused barcodes were amplified and sequenced to monitor double‐mutant abundance and to infer genetic interactions. Details of donor and recipient strain construction are shown in Figs EV1 and EV2, respectively. Media details are shown in Fig EV3.
