Figure EV1. Donor toolkit construction.
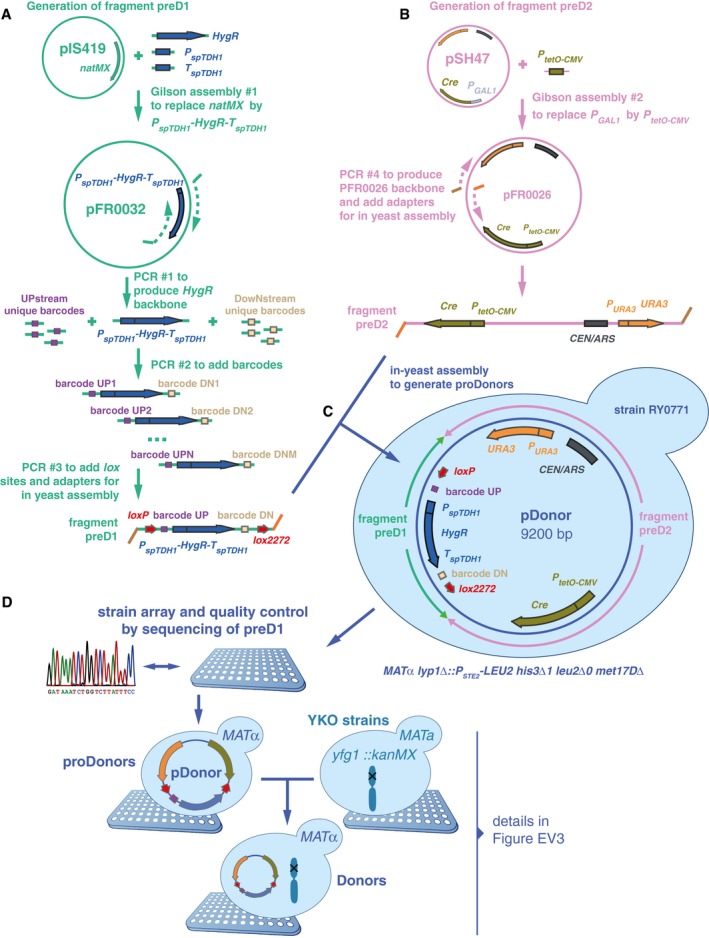
- Two fragments were built to generate proDonor plasmids. The first, preD1, contained loxP/lox2272 sites flanking two 20‐bp unique barcodes and a hygromycin resistance marker. In this study, only the upstream barcode was used for further steps, and for simplification, the downstream barcode was omitted from Fig 1.
- The second, preD2, contained the Cre recombinase driven by the doxycycline‐inducible tetO‐CMV, and a URA3 marker.
- The two fragments were assembled in vivo in yeast to generate pDonors.
- pDonors were arrayed and Sanger‐sequenced to confirm the integrity of the preD1 fragment. ProDonors with confirmed preD1 fragments were mated with YKO strains to generate strains carrying both a uniquely barcoded pDonor and a gene deletion of interest. Then, they were sporulated and the haploid MATalpha progeny were selected using the mating type maker indicated in panel (C). Details on selective media are shown in Fig EV3.
