Figure 4. Shu complex condition‐dependent genetic interactions with MAG1, SLX4, PPH3, and RAD53 .
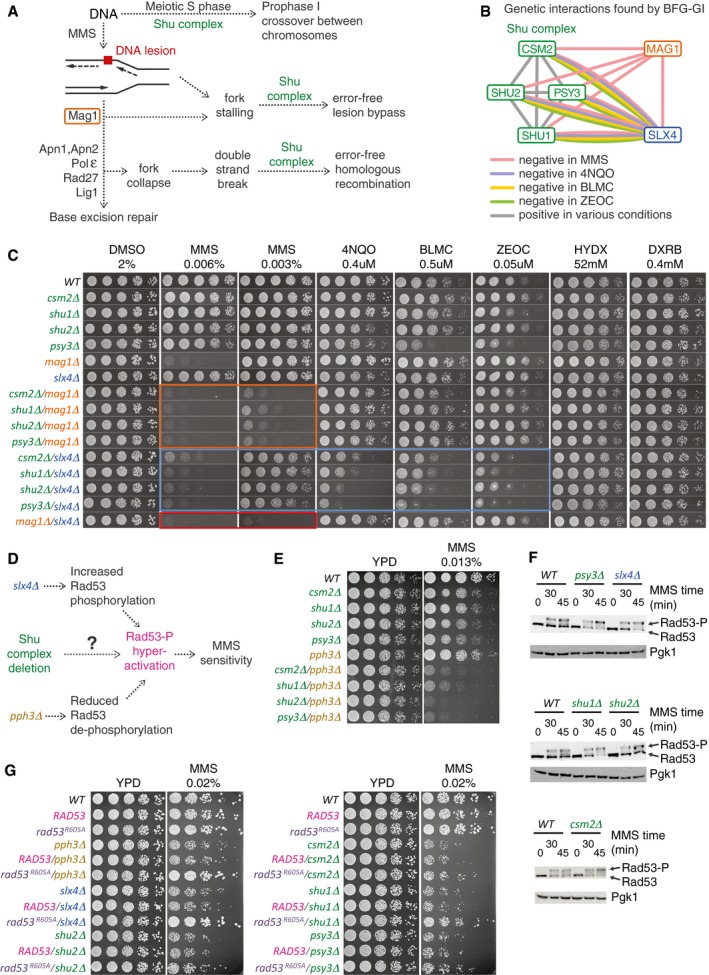
- Pleiotropic participation of the Shu complex in DNA replication and repair pathways.
- Network showing condition‐dependent genetic interactions inferred from BFG‐GI for the indicated conditions.
- Confirmation of interactions between the Shu complex, MAG1, and SLX4 using spot dilution assays including single and double mutants exposed to the indicated drugs for 48 h. Orange, blue, and red boxes indicate genetic interactions of Shu complex members with MAG1 and SLX4, and of MAG1 with SLX4, respectively.
- Schematic of potential functional connections between the Shu complex and SLX4. As with deletion of SLX4 or PPH3, deletion of Shu complex members may lead to hyperphosphorylation and hyperactivation of Rad53, resulting in increased sensitivity to MMS.
- Spot dilution assays showing genetic interactions of Shu complex genes/pph3Δ double mutants and corresponding single mutants exposed to MMS at the indicated concentration for 48 h.
- Western blot assays showing hyperphosphorylation of Rad53 in csm2Δ, psy3Δ, shu1Δ, and slx4Δ strains following treatment with 0.03% MMS. Note increased intensity of Rad53‐P bands compared with the Rad53 bands.
- Spot dilution assays of Shu complex mutants expressing a hypomorphic rad53‐R605A allele (rad53‐R605A‐6xHis‐3xFLAG‐kanMX6) compared with a wild‐type RAD53 allele (RAD53‐6xHis‐3xFLAG‐kanMX6). Cells were exposed to MMS at the indicated concentration for 60 h.
Source data are available online for this figure.
