Fig. 2.
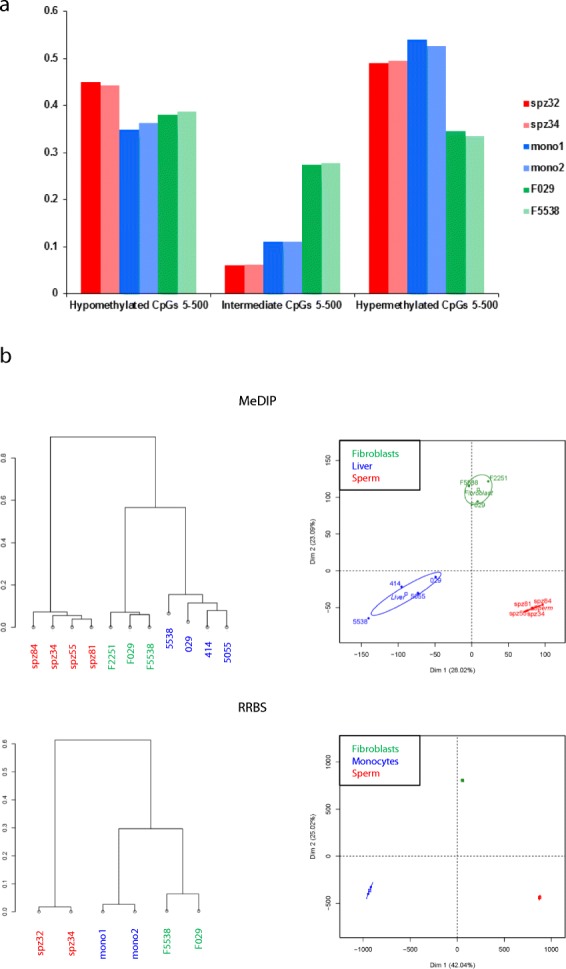
Cell type is a major determinant of DNA methylation landscapes in cattle. a Proportion of hypo- (< 20% methylation), intermediate (20–80% methylation) and hypermethylated CpGs (> 80% methylation) in each RRBS library, showing contrasted distributions between cell types. b Descriptive analyses. Upper panel: MeDIP-chip on sperm (n = 4, red), fibroblasts (n = 3, green) and liver (n = 4, blue). For each sample i and each promoter p, a normalized number of enriched probed NEpi was computed (see Additional file 1: Supplementary methods). Correlation clustering and PCA were then performed on the matrix of the normalized number of enriched probes. Lower panel: RRBS on sperm (n = 2, red), fibroblasts (n = 2, green) and monocytes (n = 2, blue). Correlation clustering and PCA were run on the totality of CpGs covered between 5 and 500 reads in the six samples
