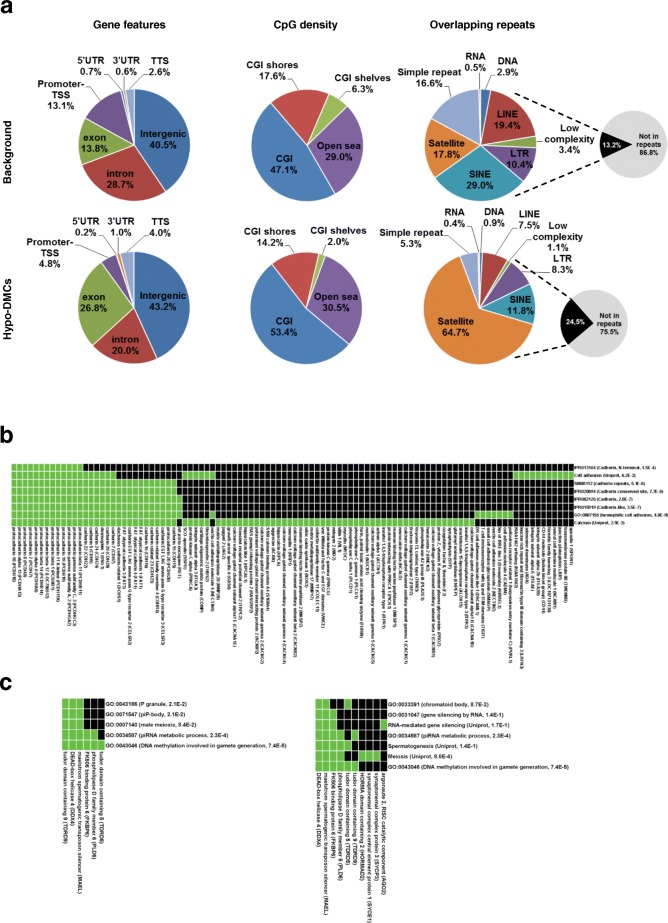
Fig. 5.
Hypo-DMCs identified by RRBS target specific genomic features and functions. The 137,861 hypo-DMCs and 1,580,644 CpGs (background) were annotated relative to gene features, CpG density and overlapping repeats. a Distribution of hypo-DMCs and background CpGs among these genomic features. b Genes with hypo-DMCs located in exons were subjected to DAVID analysis, with genes from some of the 1,580,644 CpGs in exons used as the background. The heatmap represents a functional cluster related to cell adhesion. c Genes with hypo-DMCs located in promoter-TSS were subjected to DAVID analysis, with genes containing some of the 1,580,644 CpGs in promoter-TSS used as the background. The heatmaps represent functional clusters related to piRNA metabolism (left panel) and to meiosis and spermatogenesis (right panel). To limit the size of the heatmaps, only GO terms designated as DIRECT by DAVID were used for cluster generation