Figure 1.
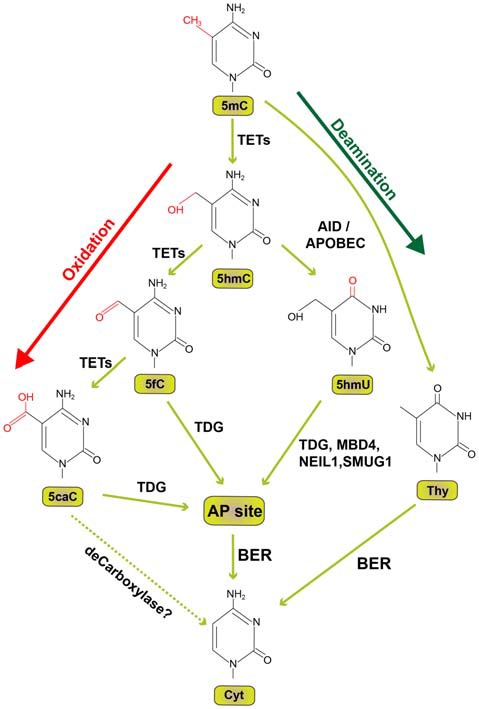
Pathways of active DNA demethylation. Since the formerly hypothesized demethylase to directly convert 5-methylcytosine (5mC) to cytosine (Cyt) has not been identified, we depict here the current view how active DNA demethylation might take place. 5mC is oxidized by ten-eleven translocation (TET) family of dioxygenases to generate 5-hydroxymethylcytosine (5hmC). In successive steps TET enzymes further hydroxylate 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Thymine DNA glycosylase (TDG) recognizes intermediate DNA forms 5fC and 5caC and excises the glycosidic bond resulting in an apyrimidinic (AP) site. In an alternative deamination pathway 5hmC can be deaminated by activity-induced cytidine deaminase/apolipoprotein B mRNA editing complex (AID/APOBEC) deaminases to form 5-hydroxymethyluracil (5hmU) or 5mC can be converted to Thymine (Thy). 5hmU can be cleaved by TDG, single-strand-selective monofunctional uracil-DNA glycosylase 1 (SMUG1), Nei-Like DNA Glycosylase 1 (NEIL1), or methyl-CpG binding protein 4 (MBD4). AP sites and T:G mismatches can be efficiently repaired by Base Excision Repair (BER) enzymes. Dotted lines indicate a proposed but not experimentally proven path.
