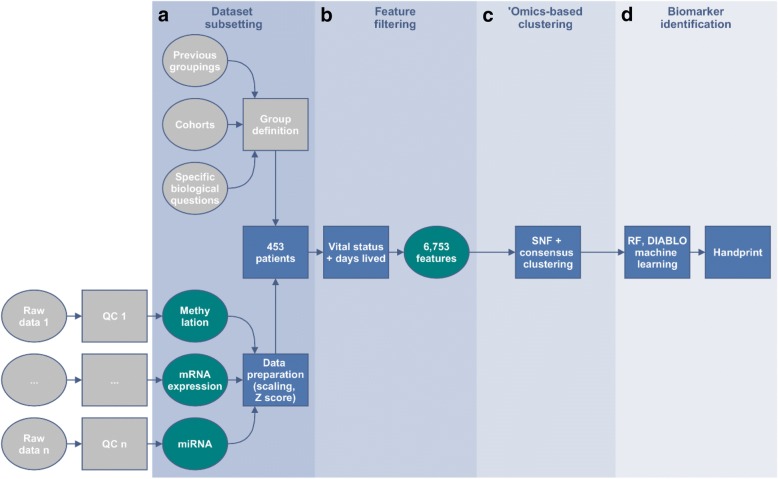
Fig. 4.
Framework outline for the TCGA handprint analysis with additional feature filtering. Each dataset was separately filtered based on nominal p-values < 0.05 when comparing alive versus deceased patients at the end of the study taking into account the total amount of days alive. A total of 6753 features were selected: 899 differentially methylated genes, 37 miRNAs and 5817 differentially expressed probesets. Consensus clustering on the fused similarity matrices determined the number of stable clusters that were viewed in a Kaplan-Meyer plot and tested for differential survival. Machine learning was then performed to identify candidate features predicting the identified groups: Recursive Feature Elimination (RFE) on a linear Support-Vector-Machine (SVM) model to identify informative features, followed by a Random Forest (RF) model building in parallel with DIABLO sPLS-DA on those features