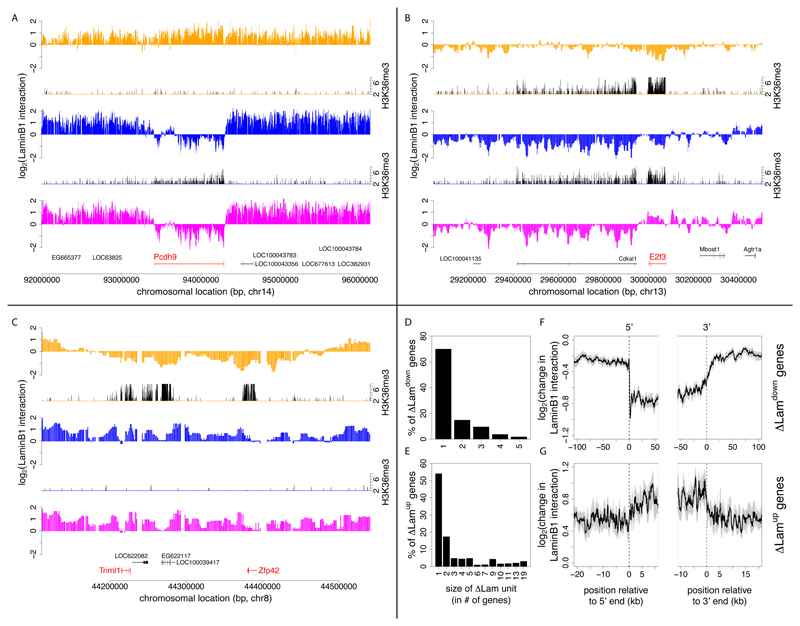
Figure 3. Changes in NL interactions involve single transcription units and multi-gene regions.
(A-C) Examples of genes that change NL interaction levels during differentiation. The log2 transformed Dam-LmnB1 over Dam methylation ratios are plotted for selected loci in ESCs (orange), NPCs (blue) and ACs (magenta). Corresponding H3K36me3 maps are shown for ESCs and NPCs (plotted in black, on orange respectively blue baselines) to indicate local transcription activity. (A) Neuron/glia-specific gene Pcdh9; (B) cell-cycle regulating gene E2f3; (C) stem cell marker genes Triml1 and Zfp42. H3K36me3 data was taken from (Mikkelsen et al., 2007). (D-E) Size distribution of relocating units during ESC→NPC transition, calculated as the number of neighboring genes with concordant significant decreases (D) or increases (E) in NL interaction levels. (F, G) Average profiles of the change in LaminB1-interaction along singleton ΔLamdown and ΔLamup genes. (F, G) Grey areas mark estimated 95% confidence intervals, the cut-offs on the x-axes are based on half the median gene size of the genes in each group.