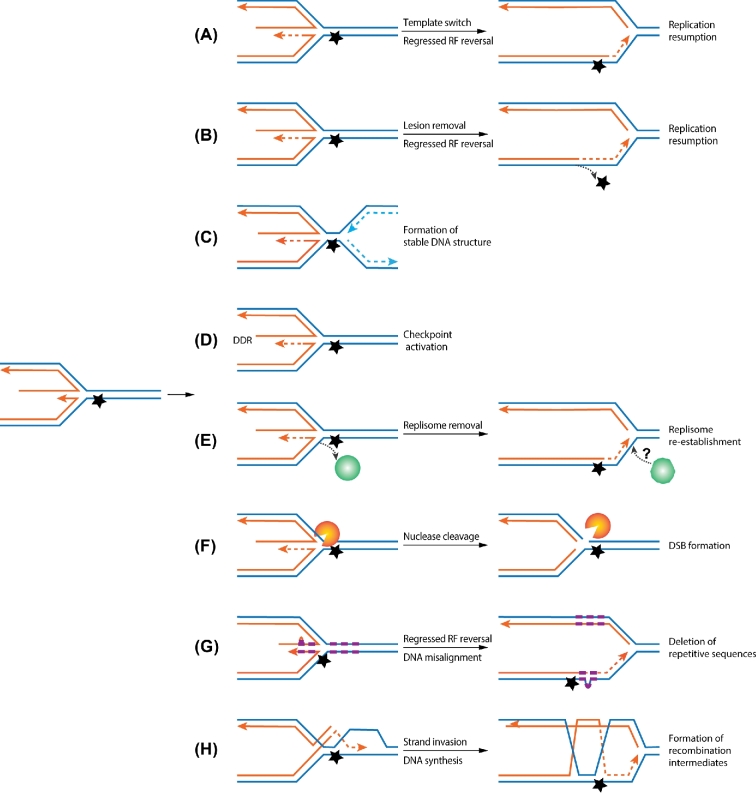
Figure 2.
Replication fork regression can lead to different molecular outcomes. Symbols and schematic features are the same as in Fig. 1. (A) Upon replication fork (RF) regression, the nascent leading strand can use the nascent lagging strand DNA as template to synthesize a stretch of DNA, thus bypassing the lesions on the parental strand. Subsequent reversal of the regression forks allows the replication fork to resume DNA synthesis. (B) Regression of replication forks can generate dsDNA regions on templates that include the region containing the damaged DNA. Changing from ssDNA to dsDNA strand forms allows the damaged DNA to be repaired using the other strand as template by the classical DNA repair pathways. (C) Replication fork regression may generate stable DNA structure, thus preventing DNA degradation and providing time for the adjacent forks to converge. (D) As dsDNA ends can be generated during replication fork regression, they can elicit the DNA damage checkpoint responses (DDR) that can induce beneficial consequences in coping with genome stress. (E) Replication fork regression can lead to eviction of replisome or its components from replication forks. This may generate the need to reload replication machineries for continued DNA synthesis. (F) Replication fork regression can lead to DSBs when the four-way ‘chicken foot’ DNA structure is cleaved by structure-specific DNA nucleases, such as the Mus81-Mms4 complex. (G) When fork regression occurs at repetitive sequences, misalignment could occur, and deletion or expansion of the DNA repeats can be produced. (H) The juxtaposition of homologous nascent and template strands as a consequence of replication fork regression can generate opportunity for recombination. One scenario is depicted where a nascent strand invades the parental strands to form recombination intermediates, which can be deleterious if not resolved.