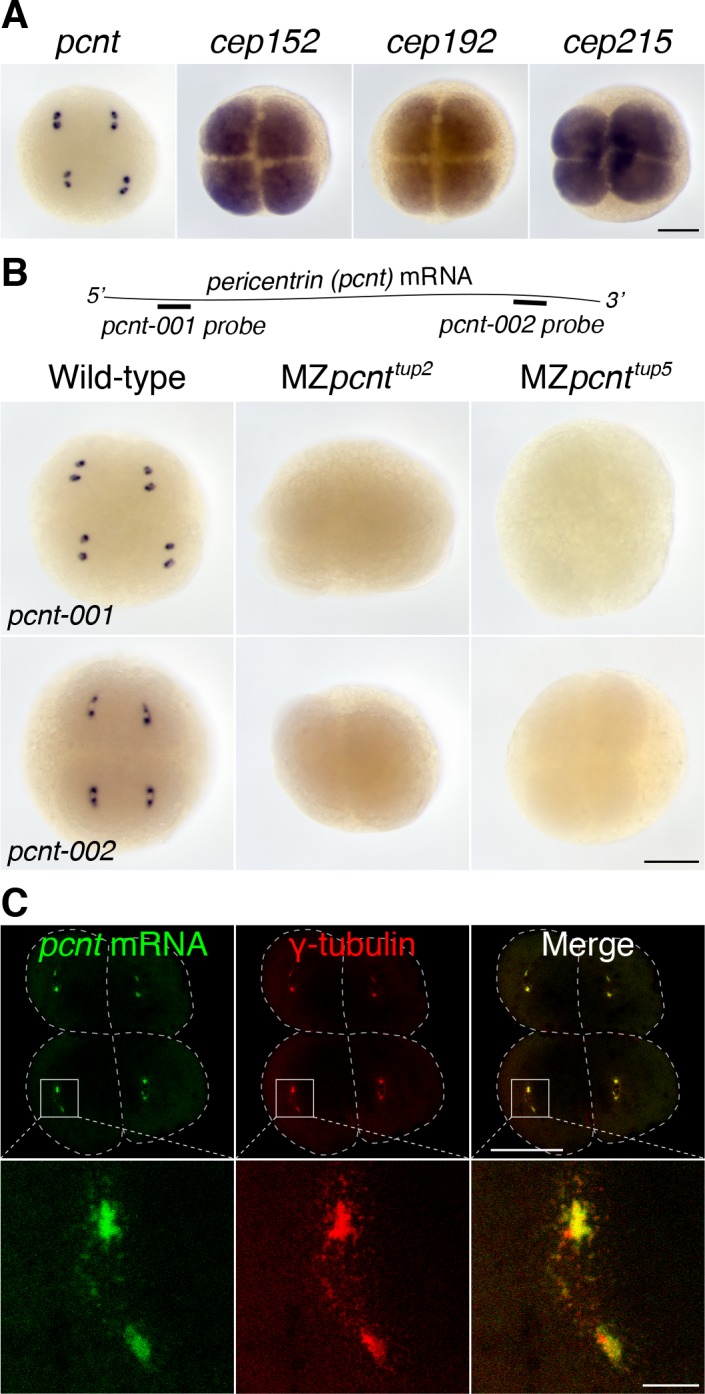
Figure 1. Pericentrin (pcnt) mRNA is localized to centrosomes in early zebrafish embryos.
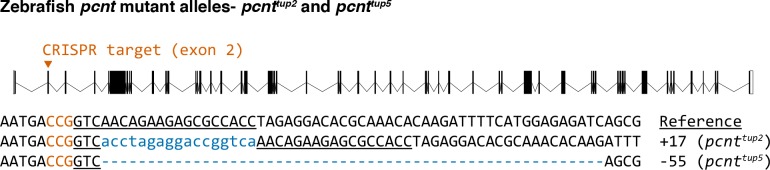
(A) RNA in situ hybridization of transcripts of different PCM components in four-cell stage zebrafish embryos. Note that while the mRNA of cep152, cep192, and cep215 displayed a pan-cellular distribution, pcnt mRNA was concentrated at two distinct foci in each cell. (B) RNA in situ hybridization showed similar dot-like patterns of pcnt transcripts with two non-overlapping antisense probes. The signals were lost in two maternal-zygotic (MZ) pcnt mutants. (C) Fluorescent RNA in situ hybridization and anti-γ-tubulin co-staining demonstrated the centrosomal localization of pcnt mRNA. n > 300 (pcnt-001 probe), n > 100 (pcnt-002 probe), n > 50 (cep152, cep192, or cep215 probe); all the embryos showed the same RNA distribution patterns as shown in the representative images. More than 100 MZpcnttup2 or MZpcnttup5 embryos were examined; none of them showed visible pcnt RNA in situ signals. Embryos were examined between 2- and 16-cell stages with representative four-cell stage embryos shown. Dashed lines delineate the cell boundaries. Scale bars: 200 µm or 25 µm (inset in C).