Figure 2.
Characterization of the RNA/DNA Hybrid Interactome
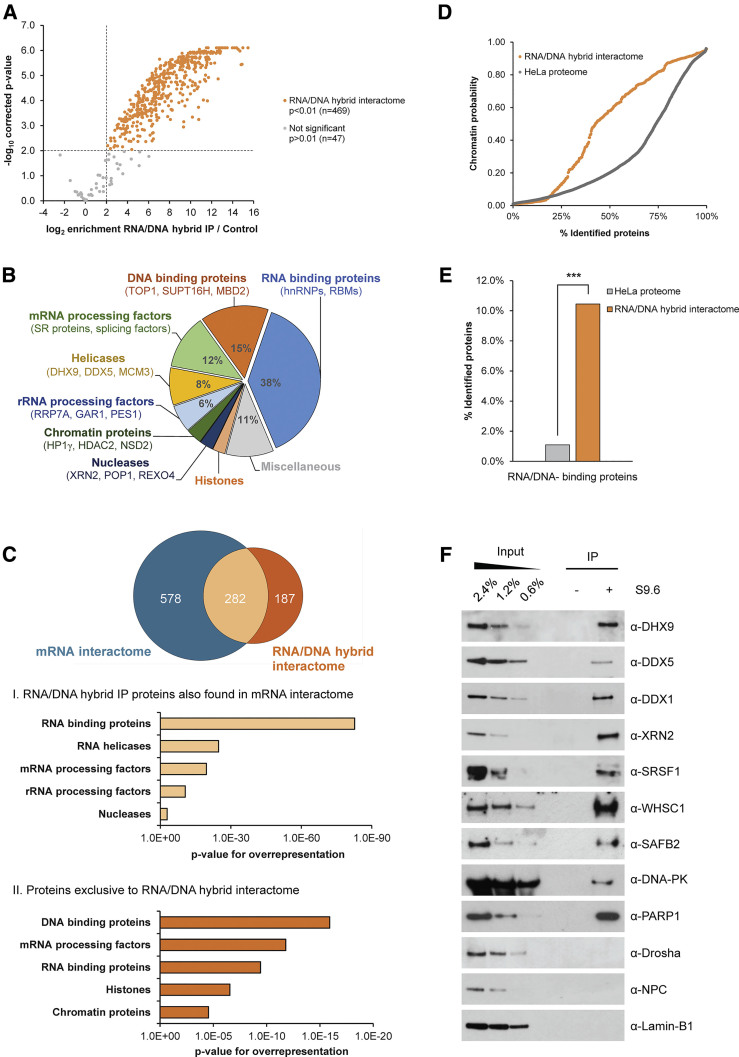
(A) Volcano plot displaying MS results of three biological replicates of RNA/DNA hybrid IP. Averaged log2 ratios between RNA/DNA hybrid IP and control IP (with addition of 1.3 μM synthetic RNA/DNA hybrid competitor) are plotted against their Benjamini-Hochberg-corrected −log10 p values across all replicates using a moderated t test. Proteins significantly enriched in RNA/DNA hybrid IP are in orange (p < 0.01) and constitute the RNA/DNA hybrid interactome. Proteins identified with p > 0.01 are in gray. Dashed lines indicate the significance cutoffs (log2 enrichment > 2 and –log10 > 2).
(B) Protein classes overrepresented in RNA/DNA hybrid interactome (corrected p < 0.05, Fisher’s exact test). Representative proteins are given in brackets.
(C) Top: overlap between mRNA interactome and RNA/DNA hybrid interactome in HeLa cells. Bottom: common proteins enriched in both RNA/DNA hybrid and mRNA interactomes (I) and proteins unique to the RNA/DNA hybrid interactome (II). The x axis indicates statistical significance of overrepresentation.
(D) The chromatin probability analysis of 7,635 HeLa proteins (HeLa proteome) and RNA/DNA hybrid interactome.
(E) Enrichment for proteins known to bind both RNA and DNA in the RNA/DNA hybrid interactome compared to the HeLa proteome (p < 1.6 × 10−28, Fisher’s exact test).
(F) Validation of RNA/DNA hybrid interactors using western blot, probed with indicated antibodies. Drosha, NPC, and Lamin B1 are negative controls. (−) IP lane corresponds to control IP with IgG2a antibody.
See also Figure S2.