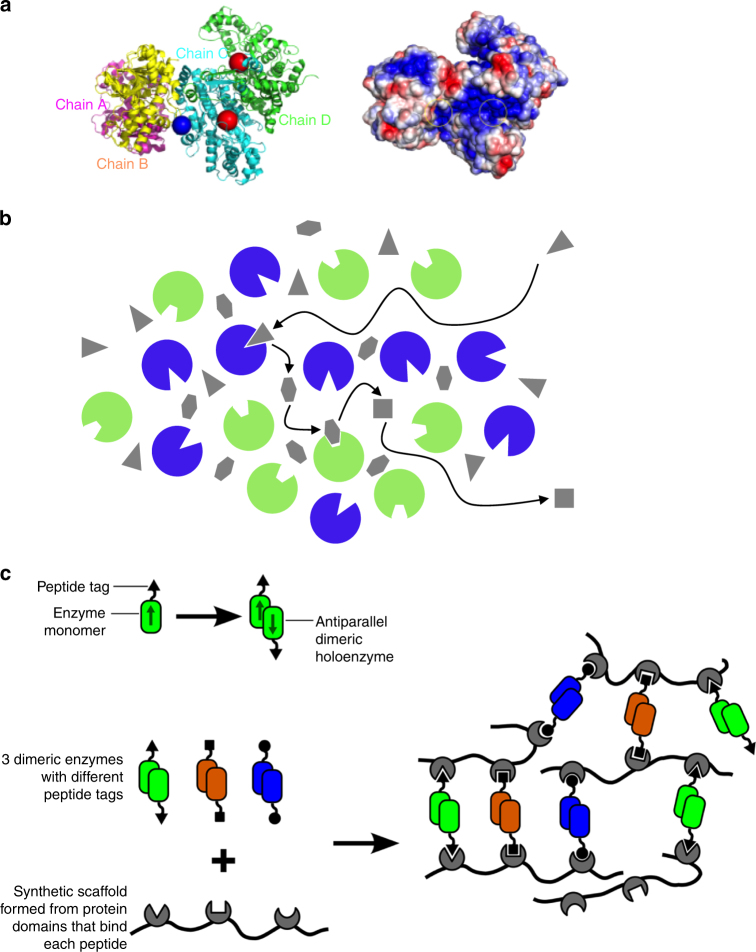
Fig. 2.
Mechanisms of substrate channelling in dynamic enzyme assemblies. a Direct channelling by electrostatic retention of the channelled metabolite on the surface of the enzyme complex. A structural model of the bovine malate dehydrogenase (MDH)–citrate synthase (CS) complex is shown. On the left, the polypeptides are illustrated as ribbon diagrams, with the MDH dimer shown in magenta and yellow and the CS dimer show in green and cyan. The blue circle shows where OAA molecules were initially placed in a Brownian dynamics simulation. Red circles show the active sites in the CS dimer. On the right, the surface structure of the complex is shown, with red and blue colours representing negative and positive electrostatic potential, respectively. Neutral regions are shown in white. Yellow circles indicate the positions of the adjacent MDH and CS active sites. b Probabilistic channelling within a large cluster of enzymes. Two enzymes are shown as green and blue circles. Metabolites are shown as grey polygons, with each shape representing a different metabolite. The arrows indicate the path taken by metabolites in a sequential conversion event by two enzymes. c Mechanism of enzyme cluster formation in synthetic scaffold-enzyme assemblies. When oligomeric enzymes are docked onto synthetic protein scaffolds via peptide tags, then interaction with more than one scaffold molecule are possible leading to the formation of a large aggregation of scaffolded enzyme complexes