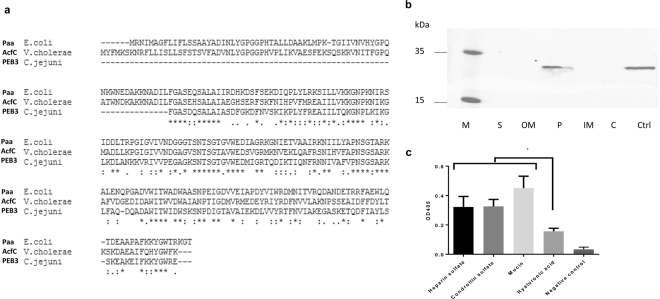
Figure 1.
(a) Blast alignments of V. cholerae AcfC with similar proteins from enteropathogenic bacteria (Paa from E. coli and PEB3 from C. jejuni) using Clustal W software; (b) Detection of V. cholerae AcfC expression in the different bacterial cell compartments using anti-AcfC antibodies. M: Page ruler plus pre-stained protein ladder (Thermo Fisher Scientific); S: Supernatant fraction; OM: Outer membrane fraction; P: Periplasmic fraction; IM: Inner membrane fraction; C: Cytoplasmic fraction; Ctrl: Ni-NTA purified His6-tagged AcfC, used as a positive control; (c) ELISA analysis of binding AcfC to different sulfated glycosaminoglycans (heparin sulfate and chondroitin sulfate), non-sufated glycosaminoglycan (hyaluronic acid) and type II porcine mucin. The values are the means ± standard deviation (n = 3). *p < 0.05, as determined by Tukey’s test.