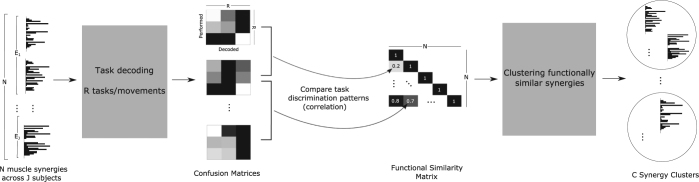
Figure 7.
Schematic illustration of our computational approach comprising three steps: decoding, functional similarity analysis and clustering. We input the single-trial activations of each of the N muscle synergies to a linear decoding algorithm that predicts which of the R movements was performed on each trial We plot the decoding results in the form of R × R confusion matrices. Each entry of the confusion matrix D(i, j) represents the fraction of trials on which movement i was decoded as j. Then we compare the confusion matrices of all synergies by computing their pairwise correlations and summarizing them in a N × N lower triangular functional similarity matrix. Finally, we use a hierarchical clustering algorithm to group functionally similar synergies onto C distinct clusters.