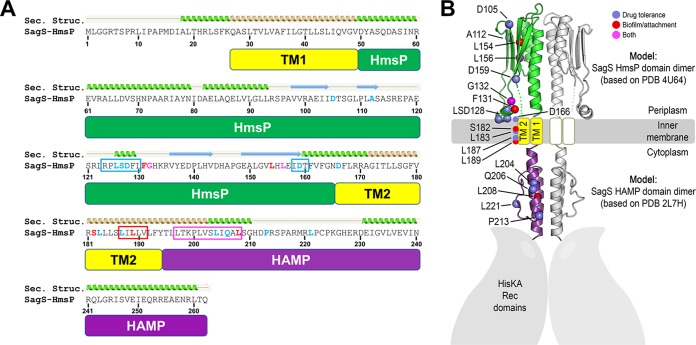
FIG 8 .
Secondary structure prediction of the HmsP and HAMP domains of SagS and spatial mapping of residues affecting SagS function. (A) Sequence of the sensory HmsP domain of SagS. Based on structural predictions (Sec. Structure) using the PHYRE2 server, the HAMP domain is shown in purple, transmembrane helices in yellow, and the sensory domain in green. Mutations affecting biofilm development (red), biofilm drug tolerance (blue), or both (magenta) are indicated in the alignment. Following the same color code, boxed regions highlight sequence motifs that were targeted for multialanine substitutions. (B) Domain modeling and spatial mutation mapping. Structural predictions were initially carried out using the PHYRE2 server. Homology models of domain dimers were calculated using Rosetta-based homology modeling. The HAMP domain is shown in purple, transmembrane helices in yellow, and the sensory domain in green. Amino acid residues (colored spheres marking C-α positions) that affect SagS function were mapped onto the three-dimensional models. Substitution of amino acids affecting attachment and biofilm formation are indicated in red, while substitutions of residues affecting susceptibility and drug tolerance are shown in blue. Amino acid residues affecting both are shown in magenta.