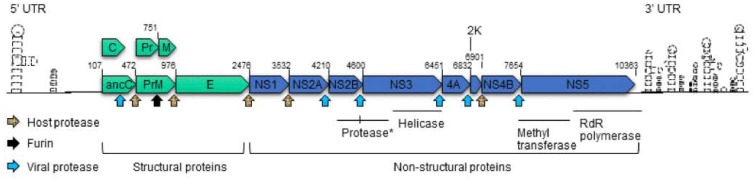
Figure 1.
Schematic structure of the Zika virus genome. Each of the viral proteins is drawn based on the relative orientation in the RNA genome. The ZIKV viral protease, host protease and Furin protease are represented by different arrows, as shown. Each arrow points to the specific protease cleavage site. The numbers shown above each protein product indicate the start/end position. Abbreviations: anaC, anchored capsid protein C; C, capsid protein C; prM, precursor membrane protein; M, membrane protein; Pr, protein pr; E, envelope protein; NS, nonstructural protein; *, protease consists of N-terminal of NS3 and C-terminal NS2B as described in the text. C-terminal of NS3 encodes helicase; 2K, signal peptide 2K; NS5 encodes methyltransferase at its N-terminal end and RNA-dependent RNA (RdR) polymerase at its C-terminal end. UTR, untranslated region. The structures of 5′ UTR and 3′ UTR are based on [21]. The information of ZIKV protein products is based on [9].