Fig. 1. Evidence for mtUPR gene activation in AD.
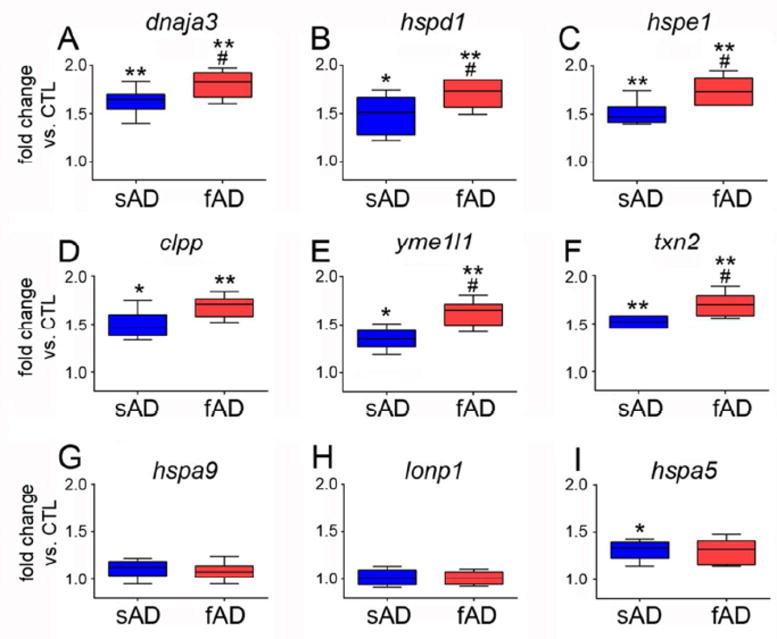
qPCR was performed on RNA extracted from frozen frontal cortex of sporadic AD (sAD) and familial AD (fAD) subjects and compared to control (CTL) subjects. Primer sets specific for mtUPR genes (A) dnaja3 (mitochondrial hsp40, or tid1), (B) hspd1 (hsp60), (C) hspe1 (hsp10), (D) clpp (ClpP mitochondrial protease), (E) yme1l1 (mitochondrial YME1-like ATPase 1), and (F) txn2 (thioredoxin 2) were compared among the groups. Primer sets specific for non-mtUPR genes (G) hspa9 (mitochondrial hsp70, or mortalin), (H) lonp1 (lon 1 mitochondrial peptidase), and (I) hspa5 (GRP78, or BiP) were also compared. Samples were run in triplicate and the ddCT method was employed to compare normalized PCR products. Box plots represent fold change in sAD and fAD compared to CTL. *p < 0.05 vs. CTL; ** p < 0.01 vs. CTL; # p < 0.05 vs. sAD.
