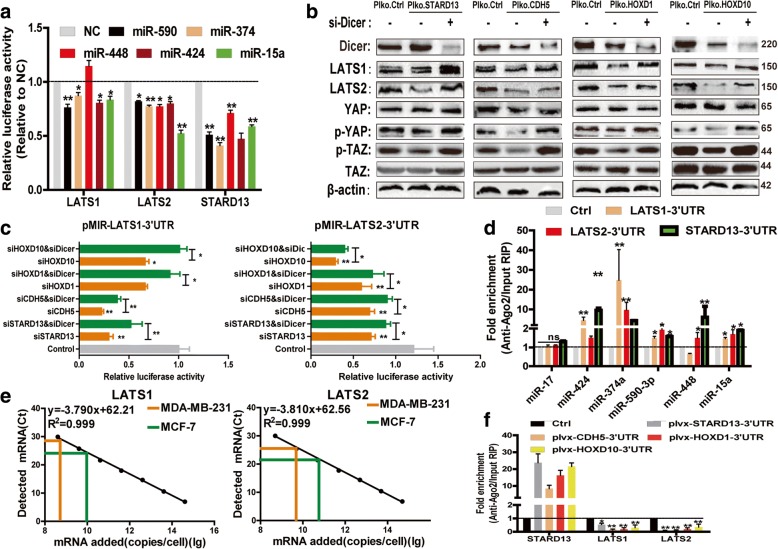
Fig. 5.
STARD13-correlated ceRNA network acts as a sub-ceRNA network to increase LATS1/2 expression. a Luciferase activity of pMIR reporter containing LATS1- or LATS2- or STARD13-3′UTR was detected upon co-transfecting with indicated miRNA mimics in 293T cells. pMIR-reporter empty vector was used as positive control. Data are presented as the ratio of luciferase activity to β-gal activity. b Western blot analysis of lysates from MCF-7 cells with STARD13-correlated ceRNA knockdown plus a control or anti-Dicer siRNA co-transfection. Depletion of Dicer rescued the expressions of LATS1/2 and downstream YAP/TAZ phosphorylation in STARD13-correlated ceRNA knocked down MCF-7 cells. c Luciferase activity of pMIR reporters which contained LATS1 (left panel) or LATS2 (right panel) 3′UTR with indicated treatment in MCF-7 cells. d qRT-PCR was used to measure the level of enrichment of miRNAs in the anti-Ago2-immunoprecipitated complexes in MCF-7 cells with empty vector or pMIR-LATS1-3′UTR or pMIR-LATS2-3′UTR or pLVX-STARD13-3′UTR transfection. miR-17 which did not hold binding sites on LATS1/2 and STARD13 was served as a negative control. e Copy number of LATS1 (left) and LATS2 (right) per cell was measured in MCF-7 cells calibrated with an internal standard curve of synthetic constructs containing LATS1 3′UTR or LATS2 3′UTR. f RIP assay of enrichment of STARD13, LATS1, and LATS2 transcripts on Ago2 relative to input in MDA-MB-231 cells with STARD13-correlated ceRNAs-3′UTR overexpression. Data were presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01 vs. control/vector