Figure 4. Paupar co‐occupies a subset of KAP1 binding sites on chromatin genome‐wide.
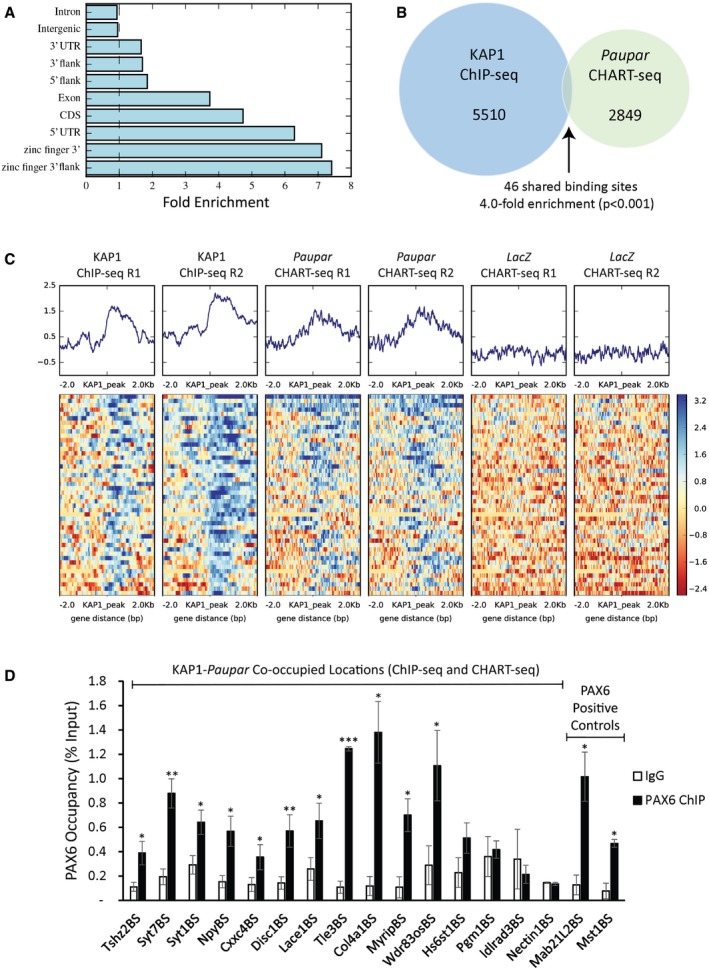
- Sites of KAP1 occupancy are particularly enriched at promoter regions (5′ UTRs), over gene bodies and over the 3′ UTR exons of zinc finger genes [q = 2 × 10−5; GAT randomisation test (Heger et al, 2013)].
- Intersection of KAP1 and Paupar binding sites in N2A cells identified 46 KAP1 bound locations that are specifically co‐occupied by Paupar. This represents a significant fourfold enrichment [P < 1 × 10−3; GAT randomisation test (Heger et al, 2013)].
- Sequencing read density distribution over the 46 shared binding locations was calculated and revealed a coincidence of Paupar and KAP1 binding site centrality. Colour legend indicates the base 2 logarithm of the ratio of read counts against input in bins of width 10 nt.
- ChIP in N2A cells was performed using either an antibody against PAX6 or an isotype‐specific control. Paupar‐KAP1 co‐occupied binding sites close to the indicated genes were amplified using qPCR to check for PAX6 chromatin association. % input was calculated as 100 × 2. Results are presented as mean values ± SEM, N = 3. One‐tailed t‐test, unequal variance *P < 0.05, **P < 0.01, ***P < 0.001.
