Figure 6. Schematic detailing possible Paupar mode of action at distal bound regulatory regions.
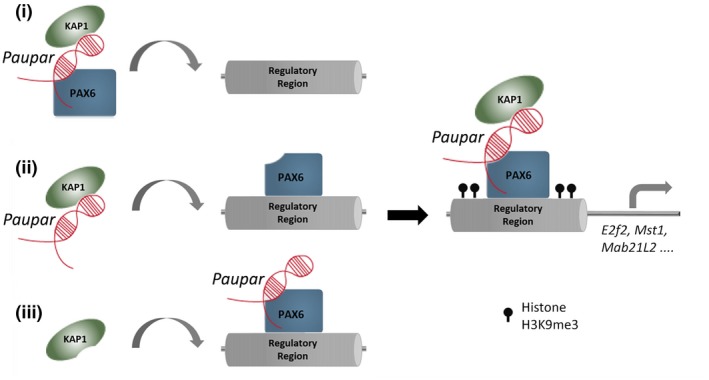
Paupar promotes KAP1 chromatin association and H3K9me3 deposition through the assembly of a DNA bound ribonucleoprotein complex containing Paupar, KAP1 and PAX6 within the regulatory regions of direct target genes such as Mab21L2, Mst1 and E2f2. We propose three potential (non‐mutually exclusive) scenarios to describe the order of assembly of this complex: (i) A ternary complex forms in the nucleoplasm before binding DNA; (ii) Paupar interacts with KAP1 and guides it to DNA bound PAX6; or (iii) KAP1 is recruited to a DNA bound PAX6‐Paupar complex. This leads to local H3K9me3 modification changes at these bound sequences in trans. The model was generated taking into consideration the discovery that Paupar genome‐wide binding sites contain an enrichment of motifs for neural transcription factors but are not enriched for sequences that are complementary to Paupar itself (Vance et al, 2014). This suggests that Paupar does not bind DNA directly but is targeted to chromatin indirectly through RNA–protein interactions with transcription factors such as PAX6. Moreover, KAP1 is a non‐DNA binding chromatin regulator that is also targeted to the genome through interactions with transcription factors.
