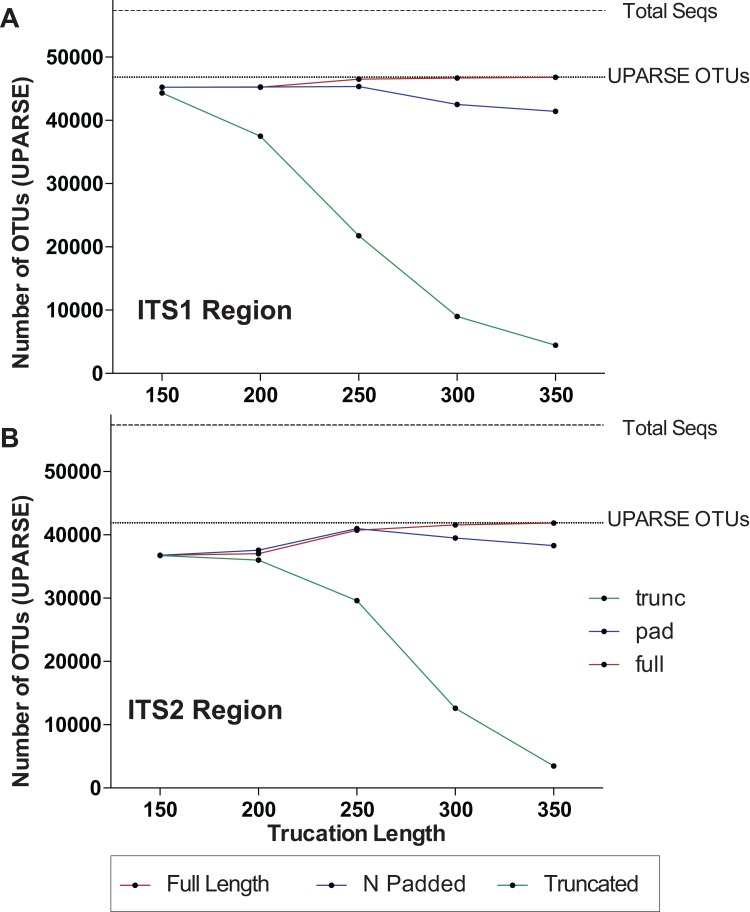
Figure 2. Pre-processing ITS sequences is critically important to accurately recover OTUs using the curated UNITE v7.2 reference database.
ITS1 and ITS2 sequences were extracted from the UNITE v7.2 general fasta release database using “AMPtk database.” Identical sequences were collapsed (dereplication) and remaining sequences were clustering using UPARSE (“cluster_otus”) to generate the total number of UPARSE OTUs expected for the (A) ITS1 and (B) ITS2 regions. The data was then processed to five different lengths (150, 200, 250, 300, and 350 bp) and then clustered (UPARSE “cluster_otus”) using (i) default UPARSE truncation (longer sequences are truncated and shorter sequences are discarded), (ii) padding with ambiguous bases (longer sequences truncated and shorter sequences padded with N’s to length threshold), and (iii) full-length sequences (longer sequences are truncated and shorter sequences are retained if reverse primer is found). Full-length and padding pre-processing sequences outperforms default UPARSE truncation.