Fig. 4.
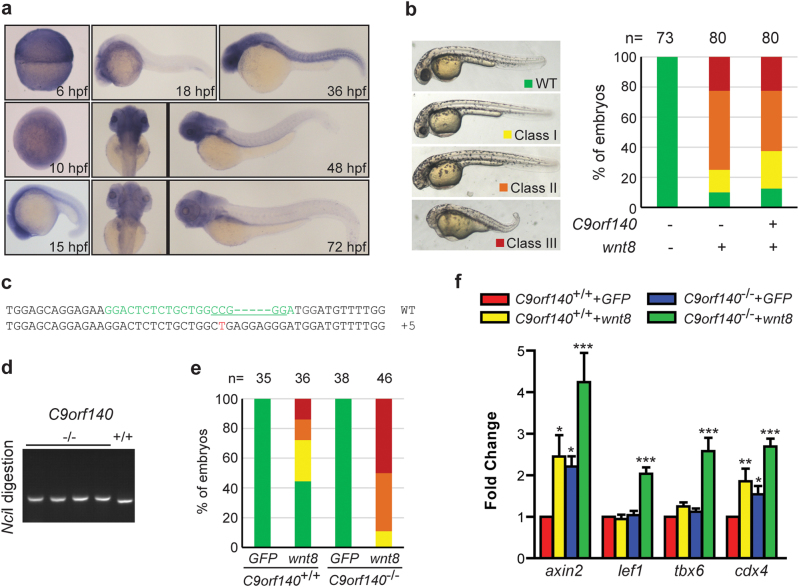
C9orf140 negatively regulates Wnt/β-catenin signaling in zebrafish. a Spatial and temporal expression of C9orf140 in zebrafish embryogenesis. The embryos at the indicated developmental stages were fixed for in situ hybridization with a C9orf140 probe. Both dorsal (left) and lateral (right) views are shown at 48 and 72 h post fertilization (hpf). b Inhibition of Wnt/β-catenin signaling by C9orf140 in zebrafish. Embryos were injected with wnt8 mRNA (5 pg) alone or together with C9orf140 mRNA (150 pg). The phenotypes of the injected embryos at 36 hpf were classified into four categories, as shown in the left panel. WT wild type; class I: small eyes; class II: headless; class III: epiboly defect or even more severe phenotype. Percentages of embryos displaying specific phenotypes are shown in the right panel. Statistical significance was calculated by Pearson χ2 test. c The sequencing results for the mutant C9orf140 allele. The CRISPR target sequence for zebrafish C9orf140 is in green and mutations in red. The NciI site used for genotype screening and identification is underlined. A 5-bp insertion in C9orf140 was identified in the C9orf140 mutant zebrafish line. d Genotypic identification of the C9orf140 mutant zebrafish lines. Genomic DNA extracted from the caudal fin was used as the template to amplify C9orf140 containing the CRISPR target sequence. The PCR products were digested by NciI and separated by electrophoresis. e Disruption of zebrafish C9orf140 facilitates Wnt signaling activation. Embryos from TU or C9orf140 -/- fish were injected with wnt8 (2 pg) or GFP (100 pg) and scored for the phenotypes shown in (b). Statistical significance was calculated by Pearson χ2 test. f The embryos injected as described in (e) were collected at the shield stage. The axin2, lef1, tbx6, and cdx4 expression levels were detected by real-time PCR and normalized to β-actin gene expression. The normalized value in the TU + GFP group was set to 1, and the values of other groups were normalized to this value. The experiment was independently performed three times, and similar results were obtained. The data shown are derived from a representative experiment expressed as the mean + SD. Statistical significance was calculated by one-way ANOVA, and significance was defined as ***p < 0.001, **p < 0.01, or *p < 0.05