Summary
Processing of double‐stranded RNA precursors into small RNAs is an essential regulator of gene expression in plant development and stress response. Small RNA processing requires the combined activity of a functionally diverse group of molecular components. However, in most of the plant species, there are insufficient mutant resources to functionally characterize each encoding gene. Here, mutations in loci encoding protein machinery involved in small RNA processing in soya bean and Medicago truncatula were generated using the CRISPR/Cas9 and TAL‐effector nuclease (TALEN) mutagenesis platforms. An efficient CRISPR/Cas9 reagent was used to create a bi‐allelic double mutant for the two soya bean paralogous Double‐stranded RNA‐binding2 (GmDrb2a and GmDrb2b) genes. These mutations, along with a CRISPR/Cas9‐generated mutation of the M. truncatula Hua enhancer1 (MtHen1) gene, were determined to be germ‐line transmissible. Furthermore, TALENs were used to generate a mutation within the soya bean Dicer‐like2 gene. CRISPR/Cas9 mutagenesis of the soya bean Dicer‐like3 gene and the GmHen1a gene was observed in the T0 generation, but these mutations failed to transmit to the T1 generation. The irregular transmission of induced mutations and the corresponding transgenes was investigated by whole‐genome sequencing to reveal a spectrum of non‐germ‐line‐targeted mutations and multiple transgene insertion events. Finally, a suite of combinatorial mutant plants were generated by combining the previously reported Gmdcl1a, Gmdcl1b and Gmdcl4b mutants with the Gmdrb2ab double mutant. Altogether, this study demonstrates the synergistic use of different genome engineering platforms to generate a collection of useful mutant plant lines for future study of small RNA processing in legume crops.
Keywords: CRISPR/Cas9, mutagenesis, soya bean, Medicago, small RNA, Drb2
Introduction
Genome engineering technologies encompass the construction and introduction of site‐specific nucleases (SSNs) into host genomes to create double‐stranded breaks (DSB) at targeted loci. The repair of the DSBs by the error‐prone nonhomologous end‐joining (NHEJ) pathway of the host organism often leads to nucleotide insertion and/or deletion (indels) events that can result in gene disruption. By taking advantage of this endogenous repair mechanism, plant biologists can generate modifications at targeted loci ranging in size from a single to several hundreds of base pairs in length, and in some cases much larger megabase pair deletions (Cermak et al., 2017; Qi et al., 2013; Voytas, 2013).
Site‐specific nucleases are synthetic enzymes with programmable DNA target specificities and include the (i) zinc‐finger nuclease (ZFN; Kim et al., 1996; Sander et al., 2011), (ii) transcription activator–like effector nuclease (TALEN; Christian et al., 2010; Miller et al., 2011) and (iii) clustered, regularly interspaced, short palindromic repeat (CRISPR)‐associated protein (Cas9) (CRISPR/Cas9) system (Cong et al., 2013; Mali et al., 2013). ZFN and TALEN‐mediated site‐directed mutagenesis has been reported in several plant species including maize (Zea mays), tobacco (Nicotiana tabacum), barley (Hordeum vulgare), soya bean (Glycine max) and Arabidopsis (Arabidopsis thaliana) (Cermak et al., 2011; Curtin et al., 2011; Li et al., 2012; Mahfouz et al., 2011; Wendt et al., 2013). CRISPR/Cas9 is the most recent platform and is comprised of the endonuclease (Cas9) and a sequence determining single‐stranded spacer guide RNA (gRNA; Bhaya et al., 2011). When introduced to plant cells via Agrobacterium‐mediated transformation, the random genome integration of the transfer DNA (T‐DNA) leads to expression of both the Cas9 endonuclease and the spacer gRNA, which when combined, generate a site‐specific DSB. Following transformation, the regenerated population of plants, the T0 generation, can be screened directly for lines harbouring mutations at the targeted locus and the heritable transmission of the introduced mutation(s) confirmed in the subsequent T1 generation of plants. Once a mutation has been heritably transmitted, it is considered stable and can be readily and reliably transmitted to subsequent generations.
Multiple reports have described the generation of somatic mutations using CRISPR/Cas9 in soya bean (Cai et al., 2015; Du et al., 2016; Jacobs et al., 2015; Lowder et al., 2015; Michno et al., 2015; Tang et al., 2016). To date however, few studies have demonstrated whole‐plant soya bean mutagenesis (Cai et al., 2017; Li et al., 2015). Here, we describe a set of plant expression vectors that can accommodate either the CRISPR/Cas9 or TALEN entry vectors to target the genes encoding the machinery proteins involved in small RNA processing in soya bean and M. truncatula (Baltes et al., 2014; Curtin et al., 2017; Zhang et al., 2013). Small RNAs are central regulators of gene expression associated with legume reproductive development, symbiotic nitrogen fixation, pathogen defence and the response to either biotic or abiotic stress (Arikit et al., 2014; Bustos‐Sanmamed et al., 2013; Li et al., 2010; Yan et al., 2015; Zhai et al., 2011). We have previously demonstrated that the use of site‐specific nucleases to generate mutations in the genes encoding the core machinery proteins of RNA silencing pathways is a highly successful tool for the study of small RNA biology in nonmodel plant species (Curtin et al., 2016). The newly generated mutant lines reported here greatly advance such resources and pave the way for future advances in legume small RNA biology research.
Results
Assessment of CRISPR/Cas9 functionality in soya bean hairy‐root tissue and TAL‐effector nuclease activity in yeast
Three binary plant expression vectors, each with a unique promoter, were converted into Gateway™ destination vectors. The promoters were selected for their constitutive and/or germ‐line expression and included the Glycine max ubiquitin (Gmubi), Arabidopsis ubiquitin10 (UBQ10) and Agrobacterium rhizogenes Root‐lociD (rolD) gene promoters (Figure 1a). To assess the functionality of each vector in soya bean, we used a previously reported set of CRISPR/Cas9 entry vectors that allow for the incorporation of the Cas9, along with a single (1x‐plex), or double 2x‐plex gRNA cassette, via a multiplex LR clonase reaction (Life Technologies, CA) (Baltes et al., 2014; Curtin et al., 2017; Figure 1b). We next constructed two single gRNAs entry cassettes that targeted the soya bean GmDcl3a gene (Glyma.04g057400; herein referred to as GmDcl3a) and carried out the cloning reaction to combine the Cas9 entry cassettes into each destination vector (Figure S1a). All six combinations of the CRISPR/Cas9 plant expression vectors were next transformed into A. rhizogenes strain K599 for the generation of transgenic plant tissue using a soya bean hairy‐root ex vitro transformation assay (Figure S2a; Taylor et al., 2006). DNA extracted from 2‐week‐old hairy‐root tissue was enriched for the mutated sequence by a restriction enzyme assay to remove nonmutated DNA sequences, and this preparation was subsequently used as PCR template (Figure S2b). A target amplicon clone and sequence (TACAS) assay confirmed deletions at both GmDcl3a loci (GmDcl3a‐1 and GmDcl3a‐2) from all three assessed expression vectors (Figure S2c).
Figure 1.
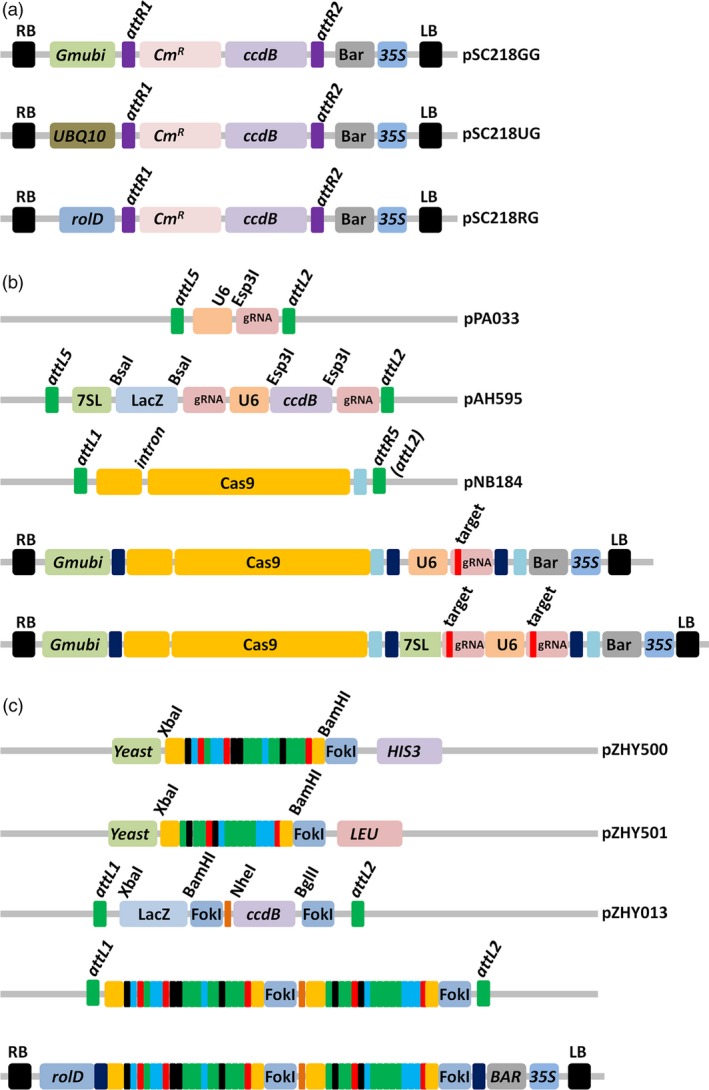
Constructs used in this study. (a) The plant expression vectors used to generate site‐specific mutations in whole‐plant soya bean: the Glycine max ubiquitin (Gmubi) promoter (pSC218GG), the Arabidopsis ubiquitin10 promoter (pSC218UG) and the Agrobacterium rhizogenes rolD promoter (pSC218RG). (b) The Cas9 and guide RNA entry vectors used for multisite Gateway™ assembly into the destination plant expression vectors. Target1 and Target2 gRNAs are cloned into the BsaI and Esp3I sites, respectively, using a golden gate ligation reaction. The ccdB and LacZ selection cassettes assist colony screening efficiency of the ligation reaction. The schematic representation of assembled reagents, a single (second from bottom) and a 2x‐plex (bottom) gRNA combined with a Gmubi‐expressed Cas9 cassette. (c) Schematic representations of the TAL‐effector assembly reagents. Target RVDs (shown as red, black, green and blue coloured rectangles) were assembled using golden gate ligation reaction into the yeast expression vectors, pZHY500 and pZHY501, for the assessment of their cleavage activity (Figure S3). Selected arrays were sequentially cloned using the depicted restriction enzymes into the pZHY013 entry vector and assembled into the plant expression vector pSC218RG by a Gateway™ reaction.
To test whether the three assessed plant expression vectors could also be used in conjunction with a previously reported TALEN platform, we selected three soya bean genes to be targeted for TALEN‐directed mutagenesis, namely GmDcl2a (Glyma.09g025400), GmDcl2b (Glyma.09g025300) and GmDcl3a (Glyma.04g057400). Furthermore, the TAL‐effector Nucleotide Targeter 2.0 (TALE‐NT) software was used to identify suitable repeat variable domains (RVD) array configurations (Doyle et al., 2012; Zhang et al., 2013), and the configured arrays were rapidly assembled using the Golden Gate TAL‐effector nuclease (TALEN) kit. The assembled arrays were next tested for their respective cleavage activities using a yeast‐based assay (Figure 1c; Method S1) (Cermak et al., 2011; Christian et al., 2010), and TAL arrays determined to have a significant activity were incorporated into the plant expression vectors (Figure S3a–b).
Successful mutagenesis and mutation transmission of the soya bean Drb2ab homoeologues
A Gmubi promoter‐driven CRISPR/Cas9 reagent with two gRNAs targeting sites within both soya bean Drb2 paralog copies was constructed (Drb2a, Glyma.12g075700, and Drb2b, Glyma.11g145900; referred to as GmDrb2a and GmDrb2b herein) (Figure 2a). The U6‐driven gRNA targeted GmDrb2a and GmDrb2b (‘Target2’), while the 7sL‐driven gRNA was designed to target four Drb genes, namely GmDrb2a and GmDrb2b and two additional Drb encoding loci (Glyma.12g172100 and Glyma.13g325600) (‘Target1’) (Figure 2b). As the 7sL promoter performs poorly in soya bean and M. truncatula, we did not expect to detect any activity associated with the 7sL‐driven gRNAs. However, the resulting reagent transgene was still introduced into soya bean via Agrobacterium‐mediated transformation and four T0 plants, WPT590‐1, WPT590‐2, WPT590‐3, and WPT590‐4, were recovered from this transformation experiment (Figure S4a–d). A PCR digestion assay was used to screen for introduced mutation and this approach showed that three of the four T0 plants returned digestion‐resistant amplicons (plants, WPT590‐1, WPT590‐2 and WPT590‐4) (Figure S4a). A heteroduplex and TACAS assay were applied to confirm the mutation status of these three T0 plant lines with the two T0 plants, WPT590‐1 and WPT590‐4, determined to harbour fully mutated alleles for both the GmDrb2a and GmDrb2b loci (Figures 2c and S4–S5), while mutated and wild‐type amplicon sequences were isolated from T0 plant, WPT590‐2 (Figures S4–S5). Sequence data generated from the TACAS assay also detected the presence of extra mutant amplicons sizes from the individual plants assessed. For example, three mutated GmDrb2a alleles (∆8‐bp, ∆7‐bp and a ∆1‐bp) were observed at high frequency across multiple leaf samples (Figures 2c and S5).
Figure 2.
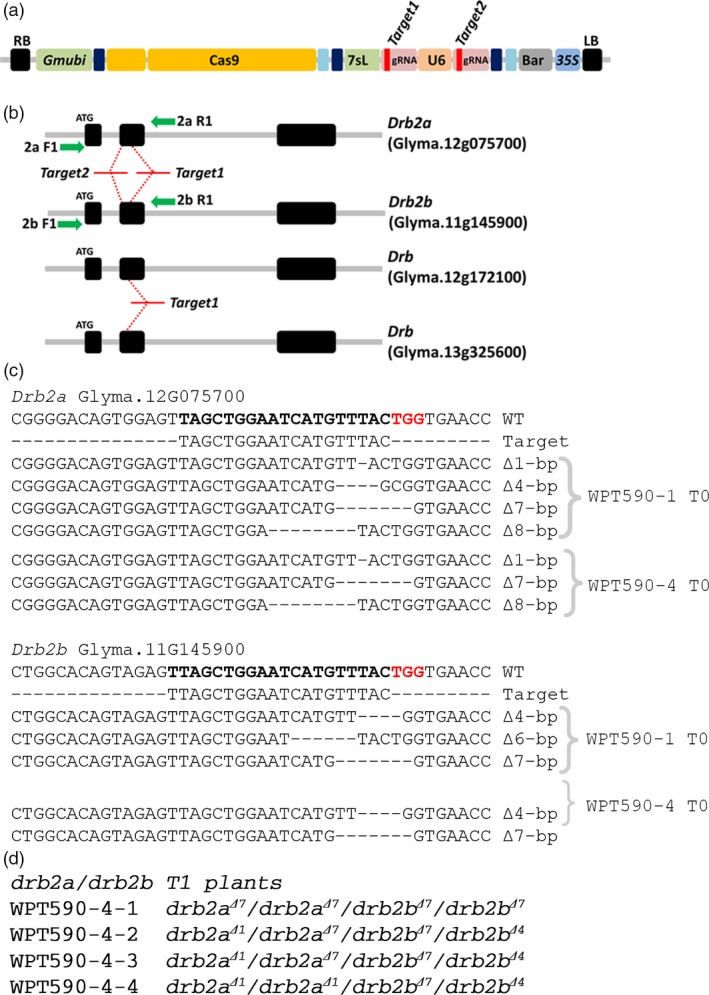
CRISPR/Cas9 mutagenesis and inheritance of mutations for the soya bean paralogous gene pair GmDrb2a and GmDrb2b. (a) Schematic representation of the CRISPR/Cas9 reagent transgene. (b) The genomic arrangement of GmDrb2a and GmDrb2b. The ‘Target1’ and ‘Target2’ represent gRNAs target sites, and the green arrows represent primer sites used to screen mutations (Table S5). (c) The multiple mutated alleles were observed in the WPT590‐1 and WPT590‐4 T0 plants from the both the TACAS and WGS analyses at the Drb2a and Drb2b loci. The target sequences are indicated in boldface, along with the PAM site (red). The ‘Target 1’ gRNA also targets two additional (although uncharacterized) Drb genes; however, no activity was observed at these sites. (d) The heritable transmission of mutated alleles was confirmed in WPT590‐1 and WPT590‐4 T1 plants. The four T1 progeny of the WPT590‐4 T0 plant shown here are examples of segregation of the Δ7‐bp and Δ1‐bp alleles for Gmdrb2a and the Δ7‐bp and Δ4‐bp alleles for Gmdrb2b.
To confirm heritable transmission of the introduced mutations, T0 plants were self‐fertilized and progeny screened (T1 plants) using the heteroduplex and TACAS approach. This approach revealed that the targeted mutations in the WPT590‐1 and WPT590‐4 T0 plants had successfully transmitted to the T1 generation (Figure S4b,d). However, this analysis also showed that no mutation or reagent transgene was transmitted to the T1 progeny of the WPT590‐2 plant (Figures S4c and S5). Whole‐genome sequencing (WGS) on the genomic DNA extracted from T0 plants, WPT590‐1 and WPT590‐4, and on two T1 progeny of each T0 plant (including T1 plants WPT590‐1‐1, WPT590‐1‐2, WPT590‐4‐1, and WPT590‐4‐2, respectively) was used to investigate the targeted loci in greater detail. The WGS approach confirmed the previously identified mutant alleles of a ∆7‐bp and ∆1‐bp for GmDrb2a and a ∆7‐bp and ∆4‐bp for GmDrb2b in both the T0 and T1 generations of the WPT590‐1 and WPT590‐4 backgrounds (Table S1; Figures S6 and S7). Interestingly, the four transmitted mutant alleles were identical between the T1 progeny derived from T0 plants WPT590‐1 and WPT590‐4. This finding indicates that the two T0 plants may have been independent regenerants of the same transformation and mutagenesis event. Furthermore, TACAS assay assessment of WPT590‐4 T1 progeny indicated that the four identified mutant alleles were segregating in the T1 generation (Figure 2d).
Upon successful heritable transmission of each mutant allele, additional somatic mutations in T1 plants are not expected to be detected due to the removal of the target sites by the bi‐allelic mutation. This was determined to be the case in these analyses, as no new mutations were identified at the targeted sites of either the GmDrb2a or GmDrb2b locus. A single wild‐type read was detected for the WPT590‐1‐1 sample; however, it is assumed likely that this read originated from contamination of the assessed T1 sample. For the T1 progeny of the T0 plant, WPT590‐4, no somatic mutations nor the presence of any wild‐type alleles was detected (Table S1). In conclusion, the WPT590‐1 and WPT590‐4 plants have fully mutated GmDrb2a (∆7‐bp and a ∆1‐bp) and GmDrb2b (∆7‐bp and a ∆4‐bp) alleles that result in the creation of in‐frame premature stop codons (Figure S8).
Identification of null‐segregant and phenotype analysis of drb2a/drb2b mutant plants
Having established the double‐mutation transmission in the T1 progeny of WPT590‐1 and WPT590‐4 T0 plants, PCR‐based assays were next used to screen greater numbers (n ≈ 50) of each T1 generation to identify null‐segregant, transgene‐free plants. However, this approach failed to identify any null‐segregant plants (Figure S4b–d), suggesting that the WPT590‐1 and WPT590‐4 backgrounds harboured three or more transgenic events. We therefore used orphaned reads (reads where only one of the two paired ends map to the transgene cassette) generated as part of our WGS analysis to identify the genomic locations of transgene insertions in T0 plants. The WGS orphaned read approach failed to identify a transgene insertion event in the WPT590‐2 background. However, this approach identified three independent transgene insertions on soya bean chromosomes four, thirteen and fifteen in the WPT590‐1 and WPT590‐4 plants (Figure 3a). Each integration site was found adjacent to small chromosomal DNA deletions (ranging from 11 to 31 bp; Figures 3a and S9–S11) and three of the flanking regions included small genomic DNA duplication or insertion events (Figure 3a). Moreover, the three identified transgene insertions were mapped to identical genomic coordinates in the WPT590‐1 and WPT590‐4 backgrounds to again indicate that these two T0 plants were independent regenerants of the same transformation and mutagenesis event.
Figure 3.
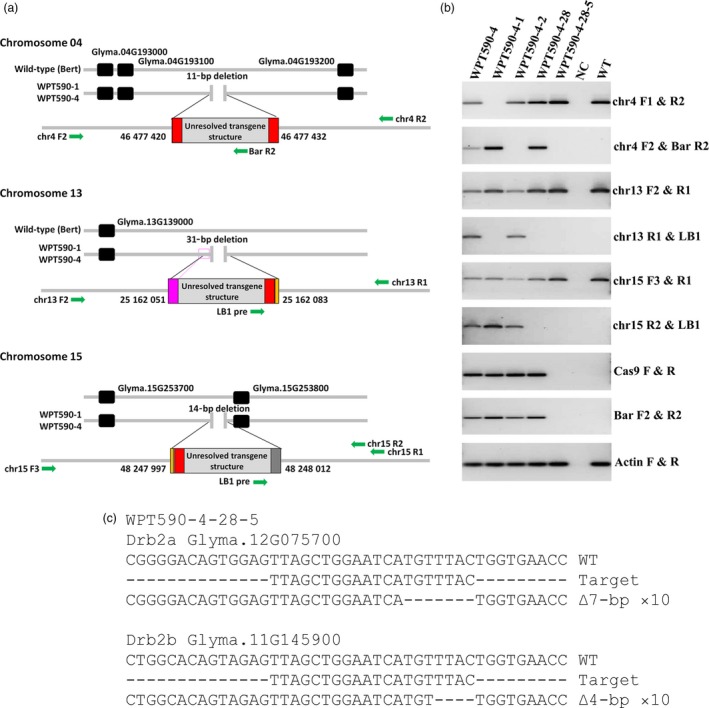
Integration sites and subsequent segregation of the CRISPR/Cas9 transgene targeting the GmDrb2 loci. (a) WGS analysis identified three transgene integrations located on chromosomes four, thirteen and fifteen, respectively. Green arrows represent primers used to screen for null‐segregants (Table S5), the pink coloured box represents a duplicated region, the orange coloured box represents the DNA insertion, and the grey coloured rectangle represents the unknown sequence that does not return a BLASTn search result to any organism. (b) A PCR assay using transgene‐specific primers and primers spanning the genomic integration sites was used to screen for the removal of reagent transgenes by genetic segregation. No amplicons could be detected from the transgene corresponding to any of three transgene genomic locations in the WPT590‐4‐28‐5 plant. (c) The WPT590‐4‐28‐5 plant was sequence‐confirmed and found to be homozygous for the drb2a Δ7 and drb2b Δ4 mutant alleles, and termed the Gmdrb2ab double mutant.
The genomic coordinates of each inserted transgene were next used to screen for null‐segregant T1 individuals and this approach revealed that T1 plant, WPT590‐4‐28, harboured only one of the three transgene insertions (Figure 3b). The applied screening also revealed that the transgene located on soya bean chromosome 4 was heterozygous (Figure 3b). Using the same approach to assess the T2 progeny of the T1 plant, WPT590‐4‐28, we identified a null‐segregant (WPT590‐4‐28‐5) that harboured no transgene‐derived sequences (Figure 3b) and confirmed via sequencing the mutant status of this plant (Figure 3c). A preliminary phenotype analysis was conducted on twenty‐four plants each of the WPT590‐4‐28 mutant (T1 progeny used in this analysis) and wild‐type ‘Bert’ backgrounds to assess the sensitivity/tolerance of the drb2ab double mutant to drought stress. Drought stress was selected as an abiotic stress of interest due to the large number of proteins associated with ‘drought stress’ or ‘osmotic stress’ responses demonstrated to have altered abundance following proteomic profiling of the Arabidopsis drb2 (Atdrb2) mutant (Reis et al., 2015b). Reis et al. (2015b) demonstrated that Atdrb2 plants were putatively more resistant to salt stress (specifically, 100 and 150 mm NaCl) than either wild‐type Arabidopsis (ecotype Columbia‐0; Col‐0), or the Arabidopsis drb1 mutant. However, we observed that 21‐day‐old Gmdrb2ab mutant plants were significantly (P = 0.01) more sensitive to drought stress than wild‐type soya bean plants when water was withheld for a 7‐day period (Figure S12a). Scanning electron microscopy was next used to analyse the adaxial and abaxial leaf surfaces of 14‐day‐old plants, but this analysis failed to detect any obvious structural defects (Figure S12c). More specifically, the stomata of Gmdrb2ab and wild‐type leaves were determined to be of a similar size and had a comparable aperture. We also quantified the abundance of several microRNA (miRNA) species in the Gmdrb2ab double mutant due to this mutant line displaying dark green leaf coloration (Figure S5), a hallmark vegetative phenotype expressed by the Atdrb2 mutant (Curtin et al., 2008; Eamens et al., 2012a). A stem‐loop primer‐based quantitative reverse transcription PCR (SL‐qPCR) approach was used to quantify the abundance of six miRNAs in the mutant and wild‐type backgrounds (Turner et al. 2013). Encouragingly, minor perturbations to the abundance of three of the six quantified miRNAs, miR156, miR163 and miR168, were detected in GmDrb2ab leaves (Figure S12b).
Successful mutagenesis of the GmDcl3a locus followed by failure to transmit the mutation and transgene
A Gmubi promoter‐driven CRISPR/Cas9 reagent with a single gRNA targeting the GmDcl3a locus was transformed into soya bean, and this transformation experiment generated five T0 plants (Figure 4a–b). Two T0 plants, WPT527‐1 and WPT527‐2, exhibited digestion‐resistant amplicons typical of germ‐line heterozygous mutations (Figure S13a). Sequencing of amplicons revealed ∆13‐bp and ∆1‐bp (~18%) and ∆4‐bp and ∆1‐bp (~12%) mutations to the GmDcl3a locus in T0 plants, WPT527‐1 and WPT527‐2, respectively (Figure 4c). Initial analysis of target site amplicons recovered mostly wild‐type alleles, rather than the expected 50% recovery of mutant amplicons normally associated with a heterozygous germ‐line mutation (Figure S14). We next screened the progeny of the WPT527‐1 and WPT527‐2 T0 plants (approximately 60 T1 plants from each T0 plant) and failed to confirm heritable transmission of any mutant allele, or the presence of the reagent transgene in either T1 population (Figure S13b). Whole‐genome sequencing was therefore next used to investigate each T0 event to determine both the fate of the mutant alleles, and to identify the genomic location of a putative reagent transgene. A comparable mutation frequency was obtained using WGS analysis to the TACAS assay for T0 plant, WPT527‐1 (17.7%) (Table S2; Figures S14 and S15) and mutations in the WPT527‐2 T0 plant were confirmed in both the GmDcl3a locus (9.1%) and the GmDcl3b pseudogene (17.8%) (Table S2; Figure S16). WGS orphaned reads identified a putative transgene location at Chr09: 36 274 816–36 275 314 for WPT527‐1; however, these same reads mapped to several genomic positions of low homology (Figure S17), and furthermore, a PCR‐based assay failed to detect any transgene sequences at the expected chromosome 9 coordinates (Figure S18). Finally, transgene analysis of the WPT527‐2 T0 plant line revealed low read counts and these lowly abundant reads failed to successfully map to any specific region of the soya bean genome.
Figure 4.
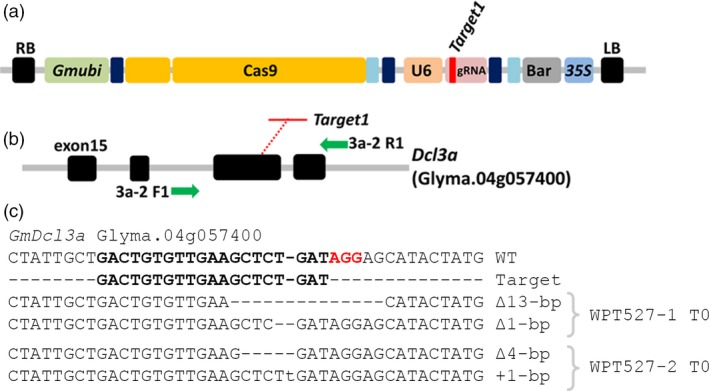
CRISPR/Cas9 mutagenesis of the soya bean GmDcl3a gene. (a) A schematic representation of the single gRNA CRISPR/Cas9 reagent targeting GmDcl3a. (b) The genomic arrangement of the GmDcl3a gene from exon 15 to exon 18, including the target site at exon 17. Green arrows represent primers used to screen mutant plants. (c) Sequence data generated from TACAS assays of the WPT527‐1 and WPT527‐2 T0 plants.
Targeted mutagenesis of the Hen1 locus in soya bean and Medicago truncatula
A Gmubi and UBQ10 promoter‐driven CRISPR/Cas9 reagent, each containing two gRNAs targeting both homoeologue copies of soya bean Hen1 locus (GmHen1a, Glyma.08g081600; GmHen1b, Glyma.05g126600) and two sites in the single copy MtHen1 gene (Medtr4g094545) were transformed into soya bean and M. truncatula (Figure 5a–b). Two T0 soya bean plants, WPT589‐1 and WPT589‐2, were screened in tissue culture for the introduction of targeted mutations using a PCR digestion assay and the WPT589‐1 plant exhibited digest‐resistant amplicons typical of a germ‐line heterozygous mutation at both the GmHen1a and GmHen1b locus (Figure S18a–d). However, this plant failed to progress through the subsequent stages of the tissue culturing process; therefore, we could not collect seed for additional analyses in the subsequent generations. The second plant, WPT589‐2, initially failed to exhibit strong evidence for germ‐line mutation. However, faint digestion‐resistant bands indicative of somatic mutations were observed in the target amplicons for both GmHen1a and GmHen1b (Figure S19a–d). The WPT589‐2 plant was recovered from tissue culture and allowed to self‐fertilize to obtain seed for further analysis. A shrunken, shrivelled seed phenotype, a phenotype similar to that expressed by the Gmdcl1a/dcl1b double mutant, was observed in approximately half of the T1 seed obtained from the WPT598‐2 T0 plant (Figure 5c). We next attempted to screen the T1 progeny of the WPT589‐2 T0 plant for introduced mutations. However, this was unsuccessful due to our inability to obtain viable seedlings from these developmentally defective seeds, as only imbibition and radical emergence were observed (Figure 5c). Intriguingly, we observed the appearance of seed mottling upon imbibition, a trait typically associated small RNA‐mediated suppression from virus infection and most recently described in soya bean plants defective in ARGONAUTE5 (AGO5) activity (Figure 5c; Cho et al., 2017; Senda et al., 2004). We next screened seeds exhibiting the mottling phenotype to identify mutations in the GmHen1a or GmHen1b loci by performing both a TACAS and heteroduplex assay across both target sites using enriched and unenriched template; however, no evidence of mutagenesis was observed (Figure S19c–d).
Figure 5.
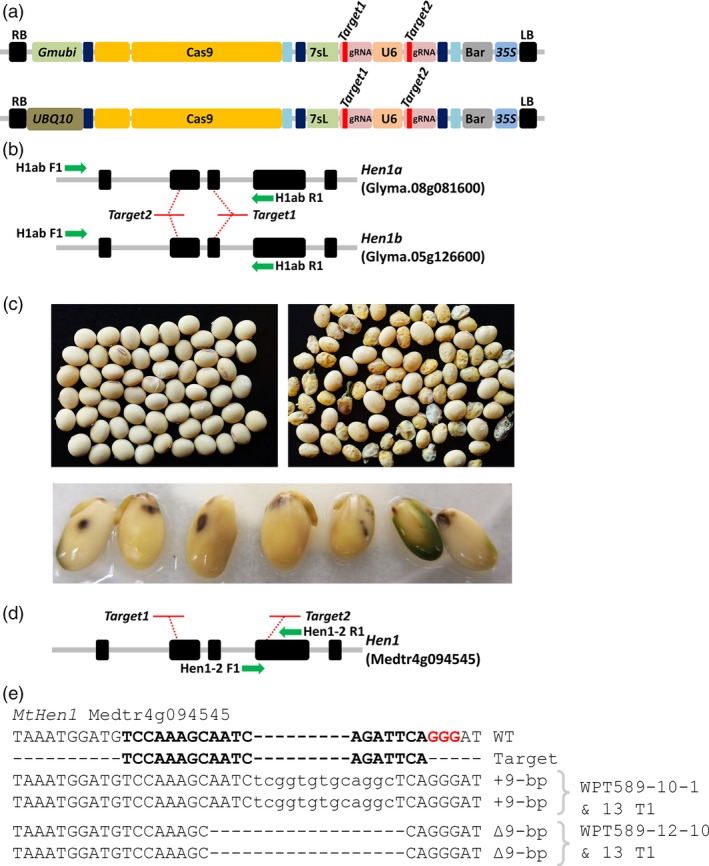
CRISPR/Cas9 mutagenesis of the soya bean and Medicago truncatula Hen1 genes. (a) A schematic representation of the reagent transgene targeting two sites in both homeologue copies of the soya bean Hen1 genes, GmHen1a and GmHen1b. The reagent transgene targeting sites in the single copy M. truncatula Hen1 gene, MtHen1, uses the At UBQ10 promoter to drive Cas9 expression. (b) The genomic arrangement of the targeted soya bean GmHen1a and GmHen1b genes and the ‘Target1’ and ‘Target2’ sites. Green arrows represent primers used to screen mutant plants (c) The phenotypes of wild type (left) and the putative Gmhen1ab double‐mutant seed (right). Seeds with the shrunken developmental phenotype germinated poorly and did not progress past the stage of radicle emergence, and also displayed a mottled phenotype (bottom panel). (d) The genomic arrangement in M. truncatula gene, MtHen1, and the location of the ‘Target1’ and ‘Target2’ sites. Green arrows represent primers used to screen mutant plants. (e) The sequence confirmation of germ‐line mutations in MtHen1 WPT222‐10 and WPT222‐12 mutant plants. The in‐frame mutations fail to disrupt the open‐reading frame of Mthen1, and no obvious hen1‐like mutant phenotype was observed.
Twelve M. truncatula T0 plants were recovered from tissue culture and four T0 plants, namely WPT222‐1, WPT222‐6, WPT222‐10 and WPT222‐12, returned digestion‐resistant amplicons indicative of germ‐line heterozygous mutations. The progeny of these four plants were screened and segregating germ‐line mutations were confirmed in the WPT222‐1, WPT222‐10 and WPT222‐12 T1 plants (Figures 5d,e and S20). Multiple WPT222‐1 and WPT222‐12 T1 plants were found to harbour an identical ∆9‐bp deletion, suggesting that these plants originated from the same transgenic event (Figure 5e). Screening of WPT222‐10 T1 plants confirmed a +9‐bp insertion (Figure 5e), and null‐segregant plants were identified from each line at a frequency that suggested each was the result of a single transgene insertion events (Figure S20).
Targeted mutagenesis of soya bean genes using TAL‐effector nuclease (TALEN)
In previous work, the soya bean dicer‐like genes GmDcl2a, GmDcl2b and GmDcl3a proved recalcitrant to mutagenesis using the zinc‐finger nuclease platform. Furthermore, and as outlined above, we also failed to generate a transmissible allele for GmDcl3a using the CRISPR/Cas9 platform. Therefore, we next attempted to use a TALEN platform to generate mutations in each of these soya bean Dcl genes. TAL arrays that showed a significant cleavage activity were cloned into a rolD expression vector (pSC218RG) and introduced into soya bean using a whole‐plant Agrobacterium‐mediated transformation assay (Table S3; Figure 6a). In total, 25 herbicide‐resistant T0 plants were recovered from tissue culture and included (i) 20 plants from the GmDcl2a TALEN transformation (plants WPT383‐1 to WPT383‐20), (ii) 4 plants from the GmDcl2b TALEN transformation (plants WPT384‐1 to WPT384‐4) and (iii) 1 plant from the GmDcl3a TALEN transformation, the WPT423‐1 T0 plant. Screening of GmDcl2a and GmDcl3a T0 plants failed to identify mutations at the targeted loci. However, three of the four T0 plants targeting Dcl2b (WPT384‐1, WPT384‐2 and WPT384‐3) exhibited digestion‐resistant amplicons indicative of a mutated target sequence (Figures 6b and S21a–b). Amplicons from the Dcl2b target site were sequenced and monoallelic deletions of ∆3‐bp for plant WPT384‐1, ∆5‐bp for plant WPT384‐2 and ∆6‐bp for plant WPT384‐3 were confirmed (Figure 6c). T0 plants were self‐fertilized, and their T1 progeny screened to confirm heritable transmission of the mutagenesis transgenes. Sequencing of the target sites in T1 progeny of the WPT384‐1 T0 plant confirmed segregation of the ∆3‐bp deletion, with six heterozygous, four homozygous and two wild‐type plants, including two transgene‐free heterozygous lines, plants WPT384‐1‐8 and WPT384‐1‐12 (Figure S22). These results confirmed the previous findings of Haun et al. (2014) and Demorest et al. (2016) that TALENs can generate nontransgenic heritable mutant plants in soya bean. A screening of T1 progeny from the T0 plants, WPT384‐2 and WPT384‐3, failed to confirm heritable transmission of the T0 mutations. Instead, screening identified mostly wild‐type genotypes and some somatic mutations (Figure S23).
Figure 6.
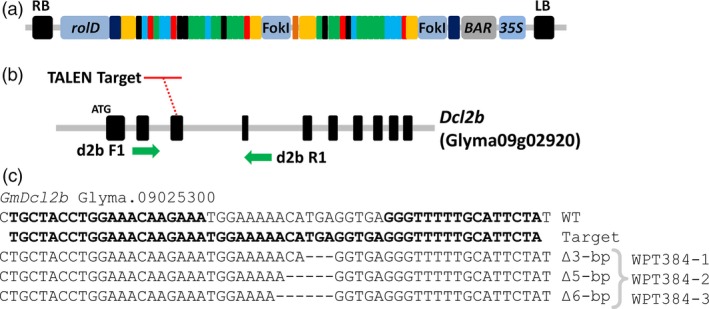
TALEN mutagenesis of the soya bean GmDcl2b gene. (a) The schematic representation of the GmDcl2b TALEN reagent expressed from the A. rhizogenes rolD promoter. (b) The genomic arrangement of GmDcl2b and the target site at the third exon. The green arrows indicate PCR primer locations used to screen mutants. (c) The sequence data from target amplicons of three T0 plants show targeted mutations. The sequences labelled as ‘Target’ indicate the 54‐bp TALEN target sequence.
Generating combinatorial soya bean mutant plants by cross‐fertilization
We have previously reported the generation of single nontransgenic Gmdcl1a, Gmdcl1b and Gmdcl4b soya bean mutants via the use of the CoDA ZFN platform (Curtin et al., 2011, 2016). Mutant alleles from two nontransgenic plants, namely DCL4a/DCL4a/dcl4b Δ2/dcl4b Δ2 and dcl1a Δ7/dcl1a Δ6/DCL1b/dcl1b Δ3, were combined by cross‐fertilization to generate a combinatorial Gmdcl1a/dcl4b double mutant (Figure S24a; Method S2). The resulting seeds were bulked, and the mutant status of F1 plants was confirmed by a PCR digestion assay that confirmed wild‐type, dcl1a Δ6 and dcl4b Δ2 allelic backgrounds (Figure S24b). The F2 progeny of these F1 plants were next screened and the homozygous double mutant was identified (Figures 7a and S24–S25). A preliminary phenotypic analysis comparing seed weight of the mutant and wild‐type plants was determined to be significantly reduced in the double mutant (P < 0.00001). These results demonstrate that the stacking of single mutants to generate a combinatorial dcl1a/dcl4b double mutant is a successful approach for the generation of novel mutant backgrounds that display unique phenotypes. We next combined the Gmdcl4b Δ2 and an additional Gmdcl1a Δ7 and Gmdcl1b Δ15 mutant alleles with the Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 plant, the Gmdrb2ab double mutant, described above (Figures S26–S30). The F1 plants resulting from this crossing event had their mutational status confirmed by a sequence‐based approach and the F2 progeny bulked for the identification of single, double and triple mutant combinations of Gmdcl1, Gmdcl1b, Gmdcl4b, Gmdrb2a and Gmdrb2b plants (Table 1).
Figure 7.
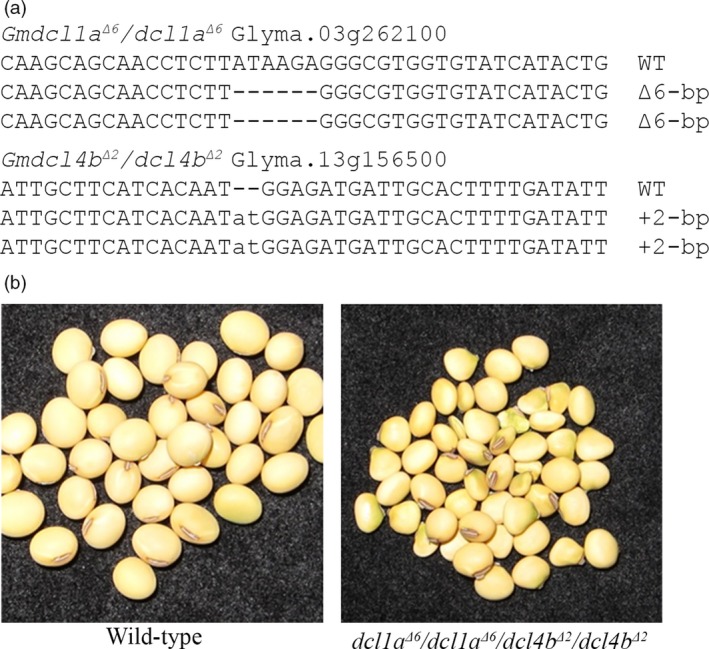
Combining soya bean dicer‐like mutated alleles. (a) Sequence confirmation of a double mutant developed via a standard genetic crossing approach using the confirmed soya bean dcl1a mutant line and the dcl4b mutant plant. (b) In a preliminary phenotype analysis, the dcl1a Δ6 /dcl1a Δ6 /dcl4b Δ2 /dcl4b Δ2 double‐mutant seed has a significantly reduced seed weight compared with wild type.
Table 1.
The generation of combinatorial dcl1ab/drb2ab and dcl4b/drb2ab mutant plants
| Reference | Direction of cross | Mutant alleles | Mutants |
|---|---|---|---|
| M13‐719 | dcl1a × dcl4b | dcl1a Δ6, dcl4b Δ2 | dcl1a/dcl4b‐1 b |
| M17‐725 | drb2ab × wt | drb2a Δ1, drb2b Δ4 | drb2a‐1, drb2b‐1 b |
| M17‐726 | wt × drb2ab | N.D. | |
| M17‐727 | drb2ab × dcl1b | drb2a Δ1, drb2b Δ4, dcl1b Δ15 | drb2a‐1, drb2b‐1 b, drb2a/dcl1b‐1 b, drb2b/dcl1b‐1 b, drb2a/drb2b/dcl1b‐1 b |
| M17‐728 | dcl1b × drb2ab | N.D. | |
| M17‐729 | drb2ab × dcl4b | drb2a Δ7, drb2b Δ7, dcl4b Δ2 | drb2a‐2, drb2b‐2 b, drb2a/dcl4b‐1 b, drb2b/dcl4b‐1 b, drb2a/drb2b/dcl4b‐1 b |
| M17‐730 | dcl4b × drb2ab a | N.D. | |
| M17‐733 | dcl1a × drb2ab a | drb2a Δ7, drb2b Δ4, dcl1a Δ7 | drb2a‐2, drb2b‐1, drb2a/dcl1a‐1, drb2b/dcl1a‐1, drb2a/drb2b/dcl1a‐1 |
| M17‐734 | dcl1a × drb2ab a | N.D. | |
| M17‐735 | dcl4b × dcl1a a | dcl1a Δ7, dcl4b Δ2 | dcl1a/dcl4b‐2 |
| M17‐736 | dcl1a × dcl4b a | N.D. |
N.D. not determined.
M1 screening in progress.
M2 screening in progress.
Discussion
Here, we report the generation of targeted mutations in loci central to the small RNA pathways of soya bean and M. truncatula using the ZFN, TALEN and CRISPR/Cas9 nuclease platforms. This approach enabled the recovery of a wide range of stable, nontransgenic plants, including those with in‐frame, frame‐shift, single homozygous and double homozygous mutations. The CRISPR/Cas9 constructs proved straightforward to produce and were efficient at generating targeted mutations in planta in both species. One of the key findings of this study was the ability of CRISPR/Cas9 reagents to generate double mutants with fully mutated alleles in the initial T0 generation, as evidenced by the GmDrb2a and GmDrb2b paralog pair. Soya bean is a highly duplicated palaeo‐polyploid plant where approximately 70% of the protein‐coding sequences encoded by the genome are further represented by a duplicated copy (Schmutz et al., 2010). An important requirement for any reagent designed for use in soya bean is the ability to simultaneously mutate multiple gene copies in a single transformation event. While the capacity of the CRISPR/Cas9 reagent to readily generate mutants is quite remarkable, we have also identified double mutants in a single generation using the ZFN platform (Curtin et al., 2016). However, ZFNs have a reduced workable target range in most plant species and require considerably more effort to design and construct (Sander et al., 2011).
We also report here on the generation of two T0 double bi‐allelic mutations of the GmDrb2a and GmDrb2b loci, and further, heritable transmission of these mutations was demonstrated in T1 progeny. The mutated alleles in these two plants were identical (this was true for both gene targets, GmDrb2a and GmDrb2b), and at least one CRISPR/Cas9 transgene insertion was transmitted to all T1 progeny. Our initial suspicion that these plants arose from the same transgenic explants was confirmed by WGS analysis that identified three transgene insertions in exactly the same genomic coordinates in both T0 plants. Using these genomic coordinates, null‐segregant transgene‐free plants were identified and deregulated article status granted by USDA‐APHIS, thereby paving the way for future field testing of this line (https://www.aphis.usda.gov/aphis/ourfocus/biotechnology/am-i-regulated) (Figure S31). The drb2ab double‐mutant plant will also be a valuable genetic resource for studying the function of DRB2 function in soya bean, having previously been demonstrated to play functionally diverse roles in multiple small RNA‐directed RNA silencing pathways in Arabidopsis, including the production stages of the repeat‐associated small‐interfering RNA (rasiRNAs) and trans‐acting small‐interfering RNAs (tasiRNAs) pathways (Pelissier et al., 2011) and the inhibition of viral replication (Barton et al., 2017). Arabidopsis DRB2 has also been shown to be an important requirement for the production of a cohort of miRNAs in developmentally important Arabidopsis tissues, namely the shoot apex and maturing pollen (Eamens et al., 2012a,b). Many of the target genes of this AtDRB2‐dependent miRNA cohort appear to be involved in stress responses and are not involved in normal plant development pathways (Reis et al., 2015b). Therefore, in view of these findings, the generation of the Gmdrb2ab double mutant provides an excellent opportunity to characterize miRNA‐directed responses to stress in soya bean and for the subsequent comparison of GmDRB2 function to the well‐documented functionality of the AtDRB2 protein.
The transgenesis and mutagenesis of the GmDcl3a locus with our CRISPR/Cas9 reagent generated curious results. First, we identified two putative T0 mutant plants that initially appeared to be germ‐line heterozygous mutants (Figure S13a). However, the quantity of mutated amplicons obtained was significantly less than that expected for a heterozygous mutant plant, which normally returns an approximate mutant to wild‐type amplicon ratio of 1 : 1 (i.e. 50% mutant and 50% wild‐type amplicons). Secondly, and most importantly, neither the targeted mutation nor the introduced transgene was heritably transmitted to the T1 generation (Figure S13b). We used the generated WGS data, specifically orphaned reads, to identify the genomic location of the transgene insertion on chromosome nine for the WPT527‐1 T0 plant. However, we failed to identify a transgene insertion for the WPT527‐2 T0 plant, despite orphaned reads mapping to the transgene at a very low frequency. There are at least three possible explanations for this finding: (i) the detected T‐DNA was present at contaminant levels; (ii) the T‐DNA was integrated into the genome; however, a stochastic process led to poor sequencing coverage; or (iii) the T‐DNA was integrated in only a small number of cells, and this cell number was insufficient for reliable detection at 20× genome coverage. Presently, insufficient data are available to make a conclusive statement on the exact reason(s) why both GmDcl3a mutations were not heritably transmitted.
We also targeted the Hen1a and Hen1b homeologue copies in soya bean and the single Hen1 copy in M. truncatula in an attempt to generate mutations in these three HEN1 encoding loci. In soya bean, we identified one plant that appeared to harbour mutations in both the Hen1a and Hen1b locus; however, this plant failed to survive through the tissue culture. A second plant was successfully recovered, and the resulting T1 progeny exhibited a seed mottling phenotype typical of small RNA perturbation. Moreover, these seeds failed to fully germinate, and we could not conclusively identify germ‐line mutations using either an enriched or nonenriched PCR template. The screening of M. truncatula T1 plants identified three mutants each with transmittable in‐frame mutations that likely do not disrupt gene function. Furthermore, none of the T1 plants exhibited the striking pleiotropic phenotypes associated with mutation of the hen1 locus in Arabidopsis and rice (Abe et al., 2010; Chen et al., 2002). Using an identical reagent platform to introduce mutations to other targeted M. truncatula genes, we have previously observed high mutagenesis rates of 50%–75% in the T0 generation plants (Curtin et al., 2017). It was therefore surprising that only in‐frame mutations were recovered at reduced mutagenesis efficiencies. Taken together, these results suggest that a heterozygous frame‐shift knockout of Hen1 may be embryo lethal in legumes.
The TALENs and CRISPR/Cas9 platforms are complementary genome engineering technologies, with the CRISPR/Cas9 platform recently emerging as the platform of choice for most applications. The TALEN platform has a limitless targeting range that might be advantageous for targeting a highly specific locus (Joung and Sander, 2013). However, the striking disadvantage of the TALEN approach, in our limited experience in soya bean, is the inferior mutational efficiency offered by this platform. Here, nine TALEN constructs that targeted three Dcl loci (GmDcl2a, GmDcl2b and GmDcl3a) were engineered. However, three of these reagents exhibited activity in yeast and just one producing a transmittable mutation. In comparison, each of the four CRISPR/Cas9 constructs used in this study showed mutation activity in whole plants and two of them produced transmittable mutations.
A combinatorial Gmdcl1a/dcl4b mutant plant was identified by combining single Gmdcl1a and Gmdcl4b mutants previously generated via the use of the ZFN platform. A similar approach has been used in Arabidopsis to establish both hierarchical and redundant roles for each of the five members of the DRB gene family in the parallel RNA silencing pathways (Curtin et al., 2008; Eamens et al., 2009, 2012a,b). We also confirmed the functional activity of GmDcl2b and GmDcl3a TALEN and CRISPR/Cas9 reagents that could be introduced into the nontransgenic Gmdcl1a/dcl4b double mutant to quickly combine additional mutations. Similarly, the Gmdrb2ab double mutant was crossed with Gmdcl1a, Gmdcl1b and Gmdcl4b single mutants, and single, double and triple mutant combinations are currently being screened. The generation of this particular suite of soya bean mutant germplasm is of high priority when Arabidopsis research is considered; that is, AtDRB2 appears to be able to bind both imperfectly and perfectly dsRNA templates, and to form functional partnerships with AtDCL1 and AtDCL4 for miRNA and siRNA (rasiRNAs and tasiRNAs) production, respectively (Eamens et al., 2012a; Pelissier et al., 2011; Reis et al., 2015a). Therefore, this collection of small RNA‐associated mutants forms a valuable genetic resource, not only for the legume research community but also for the greater plant biology community. Several recent reports have used whole‐genome, RNA and small RNA sequence analyses to document the roles played by small RNA‐directed gene expression regulation in both development and response to various environmental stresses (Arikit et al., 2014; Curtin et al., 2012; Zhai et al., 2011). To date however, small RNA studies in plant species outside of Arabidopsis have been largely limited to cataloging alterations in the accumulation of various species of small RNAs in developmentally distinct tissues, or after exposure to stress. Therefore, in legumes, as in other plant species, there has been a conspicuous lack of mutant analyses to establish gene function for loci encoding the orthologous central machinery proteins involved in either the production or activity stages of small RNA‐directed RNA silencing pathways.
Genome engineering reagents are rapidly becoming central tools for legume functional genomic studies as well as being used as an alternate approach to improve the soya bean germplasm (Cai et al., 2017; Curtin et al., 2017; Demorest et al., 2016). The limitations for targeted mutagenesis are no longer a result of inefficient genome engineering reagents. The bottleneck now appears to be the current plant transformation technologies that are highly labour intensive. Future efforts to overcome such barriers will come from the development of nontransgenic delivery methods that utilize morphogenic regulators to increase the efficiency and ease of transformation (Lowe et al., 2016; Mookkan et al., 2017). Another approach could deliver gRNAs directly to the plant that constitutively expresses Cas9 (Figure S32a–c). The ZFN, TALEN and CRISPR/Cas9 SSN platforms comprise a genome engineering toolkit that will be useful for future functional genomics projects in soya bean and in other plant species. The efficiency, multiplex capabilities and increased overall activity of the CRISPR/Cas9 platform will likely make it the preferred SSN. However, certain scenarios may arise, such as the targeting of a highly specific genomic sequence for which no CRISPR/Cas9 or ZFN target could be identified, and in this case, the TALEN platform may provide an alternate route to mutant generation.
Material and methods
Generation of plant expression vectors for site‐specific nuclease delivery
The plant expression vectors were constructed using a previously reported binary vector pNB96 (Curtin et al., 2011). A 35S::OCS cassette from pArt7 was cloned into the NotI sites of the T‐DNA alongside the 35S::BAR cassette for phosphinothricin herbicide selection (pSC218). The pSC218 vector was digested with SacII and XhoI to remove the 35S promoter and replaced with either Gmubi, AtUBQ or rolD promoters that were amplified from templates using primers with SacII and XhoI overhangs (Grefen et al., 2010; Hernandez‐Garcia et al., 2010; Wally et al., 2008). The Gateway™ reading frame cassette was removed from the pTDTO (Valdes‐Lopez et al., 2008) vectors by XhoI and SpeI digestion and cloned into the XhoI and XbaI sites to generate the pSC218GG, pSC218UG and pSC218RG vectors. The resulting plant expression vectors were used for delivery of the CRISPR/Cas9 or the TAL‐effector nuclease reagents.
Hairy‐root and whole‐plant transformation assays of soya bean and M. truncatula
Agrobacterium rhizogenes strain, K599, was used for rapid generation of transgenic plant tissue via a soya bean hairy‐root ex vitro transformation assay (Figure S2) (Curtin et al., 2011; Taylor et al., 2006). The disarmed A. rhizogenes strain, 18r12, was used for a previously reported whole‐plant transformation assay (Curtin et al., 2011; Paz et al., 2006). All whole‐plant soya bean transformations were performed in the same genetic background, the Bert cultivar. Medicago truncatula whole‐plant transformation was carried out in the cultivar, R108 (HM340), using the A. tumefaciens strain, EAH105 (Table S7; Cosson et al., 2006; Curtin et al., 2017).
CRISPR/Cas9 design and assembly
Targets sites were identified either by the ‘find’ function with the target template (G‐N20—GG) in VectoNTI (Life Technologies, CA) or were designed using the Broad Institute's sgRNA designer website (Table S4) (Doench et al., 2014). Guide RNAs for each target were engineered using a PNK oligo annealing assay and cloned into three binary vectors using Gateway™ cloning reactions. A detailed assembly method has been previously published (Curtin et al., 2017).
PCR, target amplicon cloning and the sequence (TACAS) and heteroduplex assays
All PCRs were carried out using the Hot Start Plus master mix (Qiagen) according to the manufacturer's guidelines. For the TACAS assay, PCR amplicons were purified using the QIAquick PCR Purification Kit (Qiagen) and cloned into pGem‐T‐Easy cloning vector (Promega). Colony PCR amplicons were diluted 1 : 12 with sterile deionized water and directly sequenced. The heteroduplex assay has been previously reported (Jacobs et al., 2015; Zhu et al., 2014) and can viewed online under the Method S3 section. Primers used in this report are provided in Table S5.
Analysis of transgene insertion sites and targeted and off‐target mutations
The whole‐genome sequence (WGS) analyses used to identify mutations at targeted loci and to detect the genomic location of transgene integration sites have been previously reported (Anderson et al., 2016; Srivastava et al., 2014). A more detailed description of this protocol can be found online in the Method S4. Putative off‐target mutagenesis activity in transformed plants was manually screened using WGS data, specifically via manually analysing the generated list of potential off‐target sites for each construct. The list was generated using the CRISPR‐P site (http://cbi.hzau.edu.cn/crispr/) (Table S6; Lei et al., 2014).
Phenotypic characterization of mutants
Detailed methods for characterization mutant of phenotypes that include drought assay, SL‐qPCR and SEM sample preparation can be viewed online under the Method S5.
Conflict of interest
Čermák and Voytas are named inventors on several patents involving the use of TALENs to create targeted genome modifications.
Supporting information
Figure S1 The plant expression vectors used for the delivery of CRISPR/Cas9 and TALEN site‐specific nucleases.
Figure S2 Hairy‐root transformation protocol for rapid introduction of transgenes into soybean.
Figure S3 The yeast cleavage assay used to screen TAL‐effector nuclease activity.
Figure S4 PCR‐digest and hetero‐duplex gel analysis of WPT590 soybean plants.
Figure S5 TACAS assay mutation data for WPT590‐1, WPT590‐2 and WPT590‐4 plants.
Figure S6 Whole genome sequence analysis of Drb2a (Glyma.12g075700) target sites in Bert, WPT590‐1, WPT590‐1‐1, WPT590‐1‐2, WPT590‐2, WPT590‐4, WPT590‐4‐1 and WPT590‐4‐2 plants.
Figure S7 Whole genome sequence analysis of Drb2b (Glyma.11g145900) target sites in Bert, WPT590‐1, WPT590‐1‐1, WPT590‐1‐2, WPT590‐2, WPT590‐4, WPT590‐4‐1 and WPT590‐4‐2 plants.
Figure S8 The target site encodes the start of the highly conserved second double‐stranded RNA‐binding motif (dsRBM).
Figure S9 Coverage of reads mapping to the reference genome surrounding transgene at chromosome 4.
Figure S10 Coverage of reads mapping to the reference genome surrounding transgene at chromosome 13.
Figure S11 Coverage of reads mapping to the reference genome surrounding transgene at chromosome 15.
Figure S12 Phenotype analysis of the drb2ab mutant.
Figure S13 Screening of T0 and T1 soybean plants for CRISPR‐mediated targeted mutations at Dcl3a.
Figure S14 TACAS sequence data of the Dcl3a T0 mutant plants.
Figure S15 The WGS analysis of mutations at Glyma.04g057400 in the WPT527‐1 & WPT527‐2 plants.
Figure S16 The WGS analysis of mutations at Glyma.06G05800 in the WPT527‐1 & WPT527‐2 plants.
Figure S17 WGS paired end reads mapping to the putative genomic location the Dcl3a reagent in WPT527‐1 and WPT 527‐2 T0 plant.
Figure S18 PCR assay of the putative Dcl3a reagent at the chromosome 9 locus.
Figure S19 PCR‐digestion assay of GmHen1a and GmHen1b WPT589‐1 and WPT 589‐2 T0 plants.
Figure S20 Screening of Mt Hen1 T1 mutant plants by PCR digestion assay.
Figure S21 PCR‐digestion assays of T0 plants from samples taken from different parts of the plant harboring the TALEN targeting Dcl2b plants.
Figure S22 The heritable transmission of Dcl2b mutation in WPT384‐1 T1 plants and removal of the transgene by genetic segregation.
Figure S23 The heritable transmission of the Dcl2b mutations was not observed in WPT384‐2 and WPT384‐3 T0 plants.
Figure S24 Combining dcl1a and dcl4b mutations.
Figure S25 The dcl1a Δ6 /dcl1a Δ6 /dcl4b Δ2 /dcl4b Δ2 and wild‐type plant.
Figure S26 Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 cross to wild type to recover single drb2a and drb2b mutant plants.
Figure S27 Combining the Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 and Gmdcl1b Δ15 mutant alleles.
Figure S28 Combining the Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 and Gmdcl4b Δ2 mutant alleles.
Figure S29 Combining the Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 and Gmdcl1a Δ7 mutant alleles.
Figure S30 Combining the Gmdcl4b Δ2 and Gmdcl1a Δ7 mutant alleles.
Figure S31 USDA‐APHIS confirmation that Glycine max (soybean) line WPT590‐4‐28‐5 is not a regulated article.
Figure S32 The identification of Cas9 over‐expression cassettes in whole soybean and M. truncatula plants.
Method S1 TALEN design and assembly.
Method S2 Soybean genetic hybridization assay.
Method S3 Heteroduplex assay for detection of targeted mutations.
Method S4 Identifying transgene junctions and CRIPSR deletions.
Method S5 Phenotypic characterization of mutants.
Table S1 Analysis of WGS reads form WPT590‐1, WPT590‐2 and WPT590‐4.
Table S2 Analysis of WGS reads form WPT527‐1 and WPT527‐2.
Table S3 List of TAL‐effect nuclease RVD binding arrays and gene targets.
Table S4 List of CRISPR gRNA sequences and gene targets.
Table S5 Primers used in this study.
Table S6 Potential off‐target sites generated for Dcl3a, Drb2a and Drb2b using CRISPR‐P.
Table S7 List of reagents and their targets in Medicago and soybean.
Acknowledgements
We thank Michelle Christian, Yong Zhang, Nicholas Baltes and Paul Atkins for helpful TALEN and CRISPR/Cas9 construction advice and guidance. We would also like to thank Owen Wally for the rolD promoter, Yiping Qi for the UBQ10 promoter, Gail Celio from the University Imaging Centers, University of Minnesota, for the imaging work and Diana Trujillo for photography. This work was supported by the Minnesota Research and Promotion Council (projects 501‐6120‐534 and 509‐7520‐534).
Contributor Information
Shaun J. Curtin, Email: curti242@umn.edu.
Robert M. Stupar, Email: stup0004@umn.edu.
References
- Abe, M. , Yoshikawa, T. , Nosaka, M. , Sakakibara, H. , Sato, Y. , Nagato, Y. and Itoh, J. (2010) WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining MicroRNA and trans‐acting small interfering RNA accumulation in rice. Plant Physiol. 154, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.E. , Michno, J.‐M. , Kono, T.J.Y. , Stec, A.O. , Campbell, B.W. , Curtin, S.J. and Stupar, R.M. (2016) Genomic variation and DNA repair associated with soybean transgenesis: a comparison to cultivars and mutagenized plants. BMC Biotechnol. 16, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit, S. , Xia, R. , Kakranam, A. , Huang, K. , Zhai, J. , Yan, Z. , Valdés‐López, O. , et al (2014) An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell, 26, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes, N.J. , Gil‐Humanes, J. , Cermak, T. , Atkins, P.A. and Voytas, D.F. (2014) DNA replicons for plant genome engineering. Plant Cell, 26, 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D.A. , Roovers, E.F. , Gouil, Q. , da Fonseca, G.C. , Reis, R.S. , Jackson, C. , Overall, R.L. et al (2017) Live cell imaging reveals the relocation of dsRNA binding. Mol. Plant Microbe Interact. 6, 435–443. [DOI] [PubMed] [Google Scholar]
- Bhaya, D. , Davison, M. and Barrangou, R. (2011) CRISPR‐Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. [DOI] [PubMed] [Google Scholar]
- Bustos‐Sanmamed, P. , Bazin, J. , Hartmann, C. , Crespi, M. and Lelandais‐Brière, C. (2013) Small RNA pathways and diversity in model legumes: lessons from genomics. Front. Plant Sci. 4, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y. , Chen, L. , Liu, X. , Sun, S. , Wu, C. , Jiang, B. , Han, T. et al (2015) CRISPR/Cas9‐mediated genome editing in soybean hairy roots. PLoS ONE, 10, e0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y. , Chen, L. , Liu, X. , Guo, C. , Sun, S. , Wu, C. , Jiang, B. et al (2017) CRISPR/Cas9‐mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. https://doi.org/10.1111/pbi.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak, T. , Doyle, E.L. , Christian, M. , Wang, L. , Zhang, Y. , Schmidt, C. , Baller, J.A. et al (2011) Efficient design and assembly of custom TALEN and other TAL effector‐based constructs for DNA targeting. Nucleic Acids Res. 39, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak, T. , Curtin, S. , Gil‐Humanes, J. , Cegan, R. , Starker, C.G. , Kono, T.J.Y. , Mathre, J.W. et al (2017) A multi‐purpose toolkit to enable advanced genome engineering in plants. Plant Cell, 29, 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Liu, J. , Cheng, Y. and Jia, D. (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development, 129, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y.B. , Jones, S.I. and Vodkin, L.O. (2017) Mutations in Argonaute5 illuminate epistatic interactions of the K1 and I loci leading to saddle seed color patterns in glycine max. Plant Cell, 29, 708–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, M. , Cermak, T. , Doyle, E.L. , Schmidt, C. , Zhang, F. , Hummel, A. , Bogdanove, A.J. et al (2010) Targeting DNA double‐strand breaks with TAL effector nucleases. Genetics, 186, 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L. , Ran, F. , Cox, D. , Lin, S. and Barretto, R. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, V. , Durand, P. , D'Erfurth, I. , Kondorosi, A. and Ratet, P. (2006) Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343, 115–127. [DOI] [PubMed] [Google Scholar]
- Curtin, S.J. , Watson, J.M. , Smith, N.A. , Eamens, A.L. , Blanchard, C.L. and Waterhouse, P.M. (2008) The roles of plant dsRNA‐binding proteins in RNAi‐like pathways. FEBS Lett. 582, 2753–2760. [DOI] [PubMed] [Google Scholar]
- Curtin, S.J. , Zhang, F. , Sander, J.D. , Haun, W.J. , Starker, C. , Baltes, N.J. , Reyon, D. et al (2011) Targeted mutagenesis of duplicated genes in soybean with zinc‐finger nucleases. Plant Physiol. 156, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin, S.J. , Kantar, M.B. , Yoon, H.W. , Whaley, A.M. , Schlueter, J.A. and Stupar, R.M. (2012) Co‐expression of soybean Dicer‐like genes in response to stress and development. Funct. Integr. Genomics, 12, 671–682. [DOI] [PubMed] [Google Scholar]
- Curtin, S.J. , Michno, J.‐M. , Campbell, B.W. , Gil‐Humanes, J. , Mathioni, S.M. , Hammond, R. , Gutierrez‐Gonzalez, J.J. et al (2016) MicroRNA maturation and MicroRNA target gene expression regulation are severely disrupted in soybean dicer‐like 1 double mutants. G3 Genes Genomes Genet. 6, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin, S.J. , Tiffin, P. , Guhlin, J. , Trujillo, D.I. , Burghardt, L.T. , Atkins, P. , Baltes, N.J. et al (2017) Validating genome‐wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 173, 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demorest, Z.L. , Coffman, A. , Baltes, N.J. , Stoddard, T.J. , Clasen, B.M. , Luo, S. , Retterath, A. et al (2016) Direct stacking of sequence‐specific nuclease‐induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol. 16, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G. , Hartenian, E. , Graham, D.B. , Tothova, Z. , Hegde, M. , Smith, I. , Sullender, M. et al (2014) Rational design of highly active sgRNAs for CRISPR‐Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, E.L. , Booher, N.J. , Standage, D.S. , Voytas, D.F. , Brendel, V.P. , Vandyk, J.K. and Bogdanove, A.J. (2012) TAL effector‐nucleotide targeter (TALE‐NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. , Zeng, X. , Zhao, M. , Cui, X. , Wang, Q. , Yang, H. , Cheng, H. et al (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217, 90–97. [DOI] [PubMed] [Google Scholar]
- Eamens, A.L. , Smith, N.A. , Curtin, S.J. , Wang, M.‐B. and Waterhouse, P.M. (2009) The Arabidopsis thaliana double‐stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA, 15, 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens, A.L. , Kim, K.W. , Curtin, S.J. and Waterhouse, P.M. (2012a) DRB2 is required for microRNA biogenesis in Arabidopsis thaliana . PLoS ONE, 7, e35933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens, A.L. , Wook Kim, K. and Waterhouse, P.M. (2012b) DRB2, DRB3 and DRB5 function in a non‐canonical microRNA pathway in Arabidopsis thaliana. Plant Signal. Behav. 7, 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen, C. , Donald, N. , Hashimoto, K. , Kudla, J. , Schumacher, K. and Blatt, M.R. (2010) A ubiquitin‐10 promoter‐based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64, 355–365. [DOI] [PubMed] [Google Scholar]
- Haun, W. , Coffman, A. , Clasen, B.M. , Demorest, Z.L. , Lowy, A. , Ray, E. , Retterath, A. et al (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. [DOI] [PubMed] [Google Scholar]
- Hernandez‐Garcia, C.M. , Bouchard, R.A. , Rushton, P.J. , Jones, M.L. , Chen, X. , Timko, M.P. and Finer, J.J. (2010) High level transgenic expression of soybean (Glycine max) GmERF and Gmubi gene promoters isolated by a novel promoter analysis pipeline. BMC Plant Biol. 10, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, T. , LaFayette, P. , Schmitz, R. and Parrott, W. (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung, J.K. and Sander, J.D. (2013) TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.G. , Cha, J. and Chandrasegaran, S. (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA, 93, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant, 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li, H. , Deng, Y. , Wu, T. , Subramanian, S. and Yu, O. (2010) Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 153, 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Liu, Z.‐B. , Xing, A. , Moon, B.P. , Koellhoffer, J.P. , Huang, L. , Ward, T.R. et al (2015) Cas9‐guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W. , Tang, X. , Zheng, X. , Voytas, D.F. et al (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, K. , Wu, E. , Wang, N. , Hoerster, G. , Hastings, C. , Cho, M.‐J. , Scelonge, C. et al (2016) Morphogenic regulators baby boom and wuschel improve monocot transformation. Plant Cell, 28, 1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz, M.M. , Li, L. , Shamimuzzaman, M. , Wibowo, A. , Fang, X. and Zhu, J.‐K. (2011) De novo‐engineered transcription activator‐like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double‐strand breaks. Proc. Natl Acad. Sci. USA, 108, 2623–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P. , Esvelt, K.M. and Church, G.M. (2013) Cas9 as a versatile tool for engineering biology. Nat. Methods, 10, 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michno, J.‐M. , Wang, X. , Liu, J. , Curtin, S.J. , Kono, T.J.Y. and Stupar, R.M. (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web‐tool and a modified Cas9 enzyme. GM Crops Food, 6, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.C. , Tan, S. , Qiao, G. , Barlow, K.A. , Wang, J. , Xia, D.F. , Meng, X. et al (2011) A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148. [DOI] [PubMed] [Google Scholar]
- Mookkan, M. , Nelson‐Vasilchik, K. , Hague, J. , Zhang, Z.J. and Kausch, A.P. (2017) Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 36, 1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz, M.M. , Martinez, J.C. , Kalvig, A.B. , Fonger, T.M. and Wang, K. (2006) Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium‐mediated soybean transformation. Plant Cell Rep. 25, 206–213. [DOI] [PubMed] [Google Scholar]
- Pelissier, T. , Clavel, M. , Chaparro, C. , Pouch‐Pelissier, M.‐N. , Vaucheret, H. and Deragon, J.‐M. (2011) Double‐stranded RNA binding proteins DRB2 and DRB4 have an antagonistic impact on polymerase IV‐dependent siRNA levels in Arabidopsis. RNA, 17, 1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L.S. , Larson, M.H. , Gilbert, L.A. , Doudna, J.A. , Weissman, J.S. , Arkin, A.P. and Lim, W.A. (2013) Repurposing CRISPR as an RNA‐guided platform for sequence‐specific control of gene expression. Cell, 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, R.S. , Hart‐Smith, G. , Eamens, A.L. , Wilkins, M.R. and Waterhouse, P.M. (2015a) Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat. Plants, 1, 1–6. [DOI] [PubMed] [Google Scholar]
- Reis, R.S. , Hart‐Smith, G. , Eamens, A.L. , Wilkins, M.R. and Waterhouse, P.M. (2015b) MicroRNA regulatory mechanisms play different roles in Arabidopsis. J. Proteome Res. 14, 4743–4751. [DOI] [PubMed] [Google Scholar]
- Sander, J.D. , Dahlborg, E.J. , Goodwin, M.J. , Cade, L. , Zhang, F. , Cifuentes, D. , Curtin, S.J. et al (2011) Selection‐free zinc‐finger‐nuclease engineering by context‐dependent assembly (CoDA). Nat. Methods, 8, 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. et al (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Senda, M. , Masuta, C. , Ohnishi, S. , Goto, K. , Kasai, A. , Sano, T. , Hong, J.‐S. et al (2004) Patterning of virus‐infected glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell, 16, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, A. , Philip, V.M. , Greenstein, I. , Rowe, L.B. , Barter, M. , Lutz, C. and Reinholdt, L.G. (2014) Discovery of transgene insertion sites by high throughput sequencing of mate pair libraries. BMC Genom., 15, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F. , Yang, S. , Liu, J. and Zhu, H. (2016) Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin‐like protein but not the one previously reported. Plant Physiol. 170, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, C. , Fuchs, B. , Collier, R. and Lutke, W. (2006) Generation of composite plants using Agrobacterium rhizogenes . Agrobacterium Protoc. 343, 155–167. [DOI] [PubMed] [Google Scholar]
- Turner, M. , Adhikari, S. and Subramanian, S. (2013) Optimizing stem‐loop qPCR assays through multiplexed cDNA synthesis of U6 and miRNAs. Plant Signal. Behav. 8, e24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes‐Lopez, O. , Arenas‐Huertero, C. , Ramirez, M. , Girard, L. , Sanchez, F. , Vance, C.P. , Luis Reyes, J. and Hernandez, G. (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus‐deficiency signalling in common bean roots. Plant, Cell Environ. 31, 1834–1843. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F. (2013) Plant genome engineering with sequence‐specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. [DOI] [PubMed] [Google Scholar]
- Wally, O. , Jayaraj, J. and Punja, Z.K. (2008) Comparative expression of beta‐glucuronidase with five different promoters in transgenic carrot (Daucus carota L.) root and leaf tissues. Plant Cell Rep. 27, 279–287. [DOI] [PubMed] [Google Scholar]
- Wendt, T. , Holm, P.B. , Starker, C.G. , Christian, M. , Voytas, D.F. , Brinch‐Pedersen, H. and Holme, I.B. (2013) TAL effector nucleases induce mutations at a pre‐selected location in the genome of primary barley transformants. Plant Mol. Biol. 83, 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Z. , Hossain, M.S. , Arikit, S. , Valdés‐López, O. , Zhai, J. , Wang, J. , Libault, M. et al (2015) Identification of microRNAs and their mRNA targets during soybean nodule development: functional analysis of the role of miR393j‐3p in soybean nodulation. New Phytol. 207, 748–759. [DOI] [PubMed] [Google Scholar]
- Zhai, J. , Jeong, D.‐H. , De Paoli, E. , Park, S. , Rosen, B.D. , Li, Y. , González, A.J. et al (2011) MicroRNAs as master regulators of the plant NB‐LRR defense gene family via the production of phased, trans‐acting siRNAs. Genes Dev. 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, F. , Li, X. , Baller, J.A. , Qi, Y. , Starker, C.G. , Bogdanove, A.J. et al (2013) Transcription activator‐like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Xu, Y. , Yu, S. , Lu, L. , Ding, M. , Cheng, J. , Song, G. et al (2014) An efficient genotyping method for genome‐modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 4, 6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The plant expression vectors used for the delivery of CRISPR/Cas9 and TALEN site‐specific nucleases.
Figure S2 Hairy‐root transformation protocol for rapid introduction of transgenes into soybean.
Figure S3 The yeast cleavage assay used to screen TAL‐effector nuclease activity.
Figure S4 PCR‐digest and hetero‐duplex gel analysis of WPT590 soybean plants.
Figure S5 TACAS assay mutation data for WPT590‐1, WPT590‐2 and WPT590‐4 plants.
Figure S6 Whole genome sequence analysis of Drb2a (Glyma.12g075700) target sites in Bert, WPT590‐1, WPT590‐1‐1, WPT590‐1‐2, WPT590‐2, WPT590‐4, WPT590‐4‐1 and WPT590‐4‐2 plants.
Figure S7 Whole genome sequence analysis of Drb2b (Glyma.11g145900) target sites in Bert, WPT590‐1, WPT590‐1‐1, WPT590‐1‐2, WPT590‐2, WPT590‐4, WPT590‐4‐1 and WPT590‐4‐2 plants.
Figure S8 The target site encodes the start of the highly conserved second double‐stranded RNA‐binding motif (dsRBM).
Figure S9 Coverage of reads mapping to the reference genome surrounding transgene at chromosome 4.
Figure S10 Coverage of reads mapping to the reference genome surrounding transgene at chromosome 13.
Figure S11 Coverage of reads mapping to the reference genome surrounding transgene at chromosome 15.
Figure S12 Phenotype analysis of the drb2ab mutant.
Figure S13 Screening of T0 and T1 soybean plants for CRISPR‐mediated targeted mutations at Dcl3a.
Figure S14 TACAS sequence data of the Dcl3a T0 mutant plants.
Figure S15 The WGS analysis of mutations at Glyma.04g057400 in the WPT527‐1 & WPT527‐2 plants.
Figure S16 The WGS analysis of mutations at Glyma.06G05800 in the WPT527‐1 & WPT527‐2 plants.
Figure S17 WGS paired end reads mapping to the putative genomic location the Dcl3a reagent in WPT527‐1 and WPT 527‐2 T0 plant.
Figure S18 PCR assay of the putative Dcl3a reagent at the chromosome 9 locus.
Figure S19 PCR‐digestion assay of GmHen1a and GmHen1b WPT589‐1 and WPT 589‐2 T0 plants.
Figure S20 Screening of Mt Hen1 T1 mutant plants by PCR digestion assay.
Figure S21 PCR‐digestion assays of T0 plants from samples taken from different parts of the plant harboring the TALEN targeting Dcl2b plants.
Figure S22 The heritable transmission of Dcl2b mutation in WPT384‐1 T1 plants and removal of the transgene by genetic segregation.
Figure S23 The heritable transmission of the Dcl2b mutations was not observed in WPT384‐2 and WPT384‐3 T0 plants.
Figure S24 Combining dcl1a and dcl4b mutations.
Figure S25 The dcl1a Δ6 /dcl1a Δ6 /dcl4b Δ2 /dcl4b Δ2 and wild‐type plant.
Figure S26 Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 cross to wild type to recover single drb2a and drb2b mutant plants.
Figure S27 Combining the Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 and Gmdcl1b Δ15 mutant alleles.
Figure S28 Combining the Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 and Gmdcl4b Δ2 mutant alleles.
Figure S29 Combining the Gmdrb2a Δ7 /drb2a Δ1/drb2b Δ7 /drb2b Δ4 and Gmdcl1a Δ7 mutant alleles.
Figure S30 Combining the Gmdcl4b Δ2 and Gmdcl1a Δ7 mutant alleles.
Figure S31 USDA‐APHIS confirmation that Glycine max (soybean) line WPT590‐4‐28‐5 is not a regulated article.
Figure S32 The identification of Cas9 over‐expression cassettes in whole soybean and M. truncatula plants.
Method S1 TALEN design and assembly.
Method S2 Soybean genetic hybridization assay.
Method S3 Heteroduplex assay for detection of targeted mutations.
Method S4 Identifying transgene junctions and CRIPSR deletions.
Method S5 Phenotypic characterization of mutants.
Table S1 Analysis of WGS reads form WPT590‐1, WPT590‐2 and WPT590‐4.
Table S2 Analysis of WGS reads form WPT527‐1 and WPT527‐2.
Table S3 List of TAL‐effect nuclease RVD binding arrays and gene targets.
Table S4 List of CRISPR gRNA sequences and gene targets.
Table S5 Primers used in this study.
Table S6 Potential off‐target sites generated for Dcl3a, Drb2a and Drb2b using CRISPR‐P.
Table S7 List of reagents and their targets in Medicago and soybean.


