Figure 5.
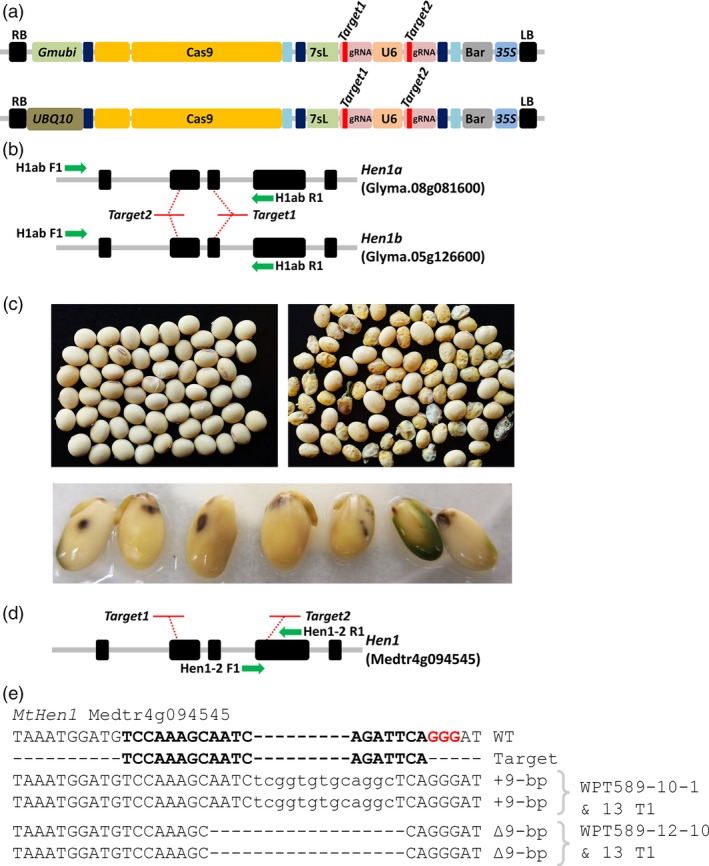
CRISPR/Cas9 mutagenesis of the soya bean and Medicago truncatula Hen1 genes. (a) A schematic representation of the reagent transgene targeting two sites in both homeologue copies of the soya bean Hen1 genes, GmHen1a and GmHen1b. The reagent transgene targeting sites in the single copy M. truncatula Hen1 gene, MtHen1, uses the At UBQ10 promoter to drive Cas9 expression. (b) The genomic arrangement of the targeted soya bean GmHen1a and GmHen1b genes and the ‘Target1’ and ‘Target2’ sites. Green arrows represent primers used to screen mutant plants (c) The phenotypes of wild type (left) and the putative Gmhen1ab double‐mutant seed (right). Seeds with the shrunken developmental phenotype germinated poorly and did not progress past the stage of radicle emergence, and also displayed a mottled phenotype (bottom panel). (d) The genomic arrangement in M. truncatula gene, MtHen1, and the location of the ‘Target1’ and ‘Target2’ sites. Green arrows represent primers used to screen mutant plants. (e) The sequence confirmation of germ‐line mutations in MtHen1 WPT222‐10 and WPT222‐12 mutant plants. The in‐frame mutations fail to disrupt the open‐reading frame of Mthen1, and no obvious hen1‐like mutant phenotype was observed.
